Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Amina Yu and Version 1 by Martin Kussmann.
Natural bioactives can be classified into micronutrients (i.e., vitamins and minerals); phytonutrients (e.g., phenolics, alkaloids, and terpenes); pre- and probiotics; and bioactive peptides. In particular, bioactive peptides have remained largely underappreciated as molecular deliverers of health promotion, mainly due to: (assumedly) poor bioavailability after oral consumption due to proteolysis along the gastrointestinal tract; limited transport from the gut lumen to the bloodstream; and, importantly, insufficient discovery and translation based on serendipitous research and/or high-throughput screening.
- bioactive
- peptide
- nutrition
- food
- plant
- ingredient
- artificial intelligence
- design
- peptidomics
- validation
- in vitro biology
1. Artificial Intelligence in (Life) Science and Technology
Human intelligence is unmatched when it comes to versatility: the human brain is extremely flexible in learning and in developing and performing a vast array of cognitive, creative, and executive tasks [11][1]. This is a major reason why humans have succeeded in populating almost every place on this planet [12][2].
However, artificial intelligence (AI) is increasingly outperforming human intelligence when it comes to data processing speed; handling huge volumes of data; establishing connections between and within large sets of unrelated pieces of information, especially at first glance; very fast learning; and—as a result—forecasting and predicting the scenarios, behaviour, and functions of complex systems [13][3].
AI is already revolutionizing various science and technology sectors. It has tremendous impact on medicine [14][4], nutrition [15][5], diagnostics [16][6], environmental science [17][7], logistics [18][8], and robotics [19][9], and this impact is growing exponentially. Rational drug design, for example, is currently greatly enhanced by AI and holds promise for yielding more efficacious drugs in a shorter time [20][10]. In medical diagnostics, AI’s superb capability of pattern recognition is widely leveraged to improve and accelerate the early and accurate detection of disease states and deviations from healthy physiology, especially when combined with imaging technologies [21][11]. The fast-learning quality of AI has enabled robots to act autonomously and react to changing conditions and situations. In environmental research on climate change, for example, AI enables the forecasting of the behaviour of hugely complex systems, such as connected biospheres and atmospheres [22][12]. Logistic companies deploy AI to design and optimize networks of transportation and communication [18][8].
AI is therefore a key to improved health care, healthier food, a more sustainable food system, and—thereby—a healthier society and planet. AI has been described to potentially reduce global healthcare costs through more efficient and more directed discovery and development of nutritional interventions [23][13]. However, in nutrition, the power of AI is only beginning to be recognized and appreciated. AI can be deployed to discover bioactive, health-beneficial peptides in natural sources [6][14]. Living systems use peptides to communicate and regulate their functions. However, when these peptides reside in plant and food proteins, they are inactive unless they are unlocked from their parent proteins. Large peptide knowledge bases lay the foundation for predicting, localizing, unlocking, and testing peptides that deliver health benefits to humans and animals and make our food healthier and more sustainable [24][15]. In other words, AI can be used to better learn how to speak the language of nature and to teach the body to maintain or restore health and improve performance.
2. Artificial Intelligence for Prediction and Discovery of Bioactive Peptides
2.1. Concept and Computation
The key strategic advantage of using AI for the prediction and discovery of bioactive peptides is that purpose definition and design can be put in front of the entire process. This contrasts with traditional serendipitous or screening approaches. By putting AI in front of the peptide discovery and development process, the path from idea and concept to solution can be shortened from decades to a few years. The number of wet laboratory experiments and pre-clinical studies, which are time- and cost-determining factors, can be substantially reduced by upfront intelligent in silico design [6][14]. First, the desired benefit is defined, be it for human or animal health or for a healthier and more sustainable food solution; then, based on interrogable peptide knowledge repositories and machine learning, bioactive peptides can be predicted to exert such benefit; such predicted peptides can then be tested in vitro. The iterative process and integrated cycle of prediction and testing, which results in computational learning, can generate a feasible number of potent bioactive candidate peptides to then be validated in vivo, in a human, or in a food technology setting. Typically, three rounds of prediction and testing are performed to produce a list of lead peptides for further ingredient development. Consequently, AI can indeed guide and empower bioactive peptide discovery, food ingredient and dietary supplement design, manufacturing, and clinical validation [6,28][14][16]. Such AI-discovered, natural bioactive peptides were recently taken from concept to pre-clinically proven solutions [29,30,31,32][17][18][19][20].
2.2. Prediction of Peptide Properties
The target benefit is defined typically around the established or new value propositions of a private company or around an established or new research area of a public research institution. In the context of this manuscript this value proposition would be either in human health care or in the food/feed system’s sustainability. The molecular mechanisms underlying the specific target benefit(s), for example, consumer/patient benefits like physical mobility, immune balance, or metabolic control, can in many cases be identified through public and proprietary knowledge mining: both public-domain ‘flat files’ like publications or patents and publicly accessible databases can be downloaded and automatically mined for relevant information, e.g., by natural language processing [33][21]. Afterwards, manual—or rather cerebral—curation is needed to ensure the quality and relevance of the insourced information. This knowledge body can be combined with proprietary databases, which contain in-house generated information on peptide properties. Through interrogating these knowledge bases, the peptides involved in the identified key molecular mechanisms and the supposedly conferred target benefit(s) can be predicted by means of AI-powered mining, which relies on predictors for peptide functions and physicochemical properties [29,34][17][22].
This can be further illustrated with the following analogy: every peptide has a unique sequence and a flexible three-dimensional (3D) structure. According to these 3D properties, every peptide can be projected into a 3D space containing all possible peptide 3D structures, thereby occupying a specific and unique position in that space. Now, the same can be envisaged with many more properties than just the 3D structure: peptides have numerous properties and various features that, when taken together, make up the entirety of individual peptide characteristics. According to these typically >100 characteristics, peptides can be projected into the high-dimensional space (representing these >100 characteristics), thereby again occupying one specific and unique position in that space. Visually speaking, this multi-dimensional space of peptides with their unique property combinations can then be “interrogated from the angle and with the depth of the desired property combination”, eventually “landing” on one or a few peptide positions that represent the best combination of the desired properties and hence the best hit(s) for a targeted, multi-faceted benefit [35][23]. This is illustrated in Figure 1.
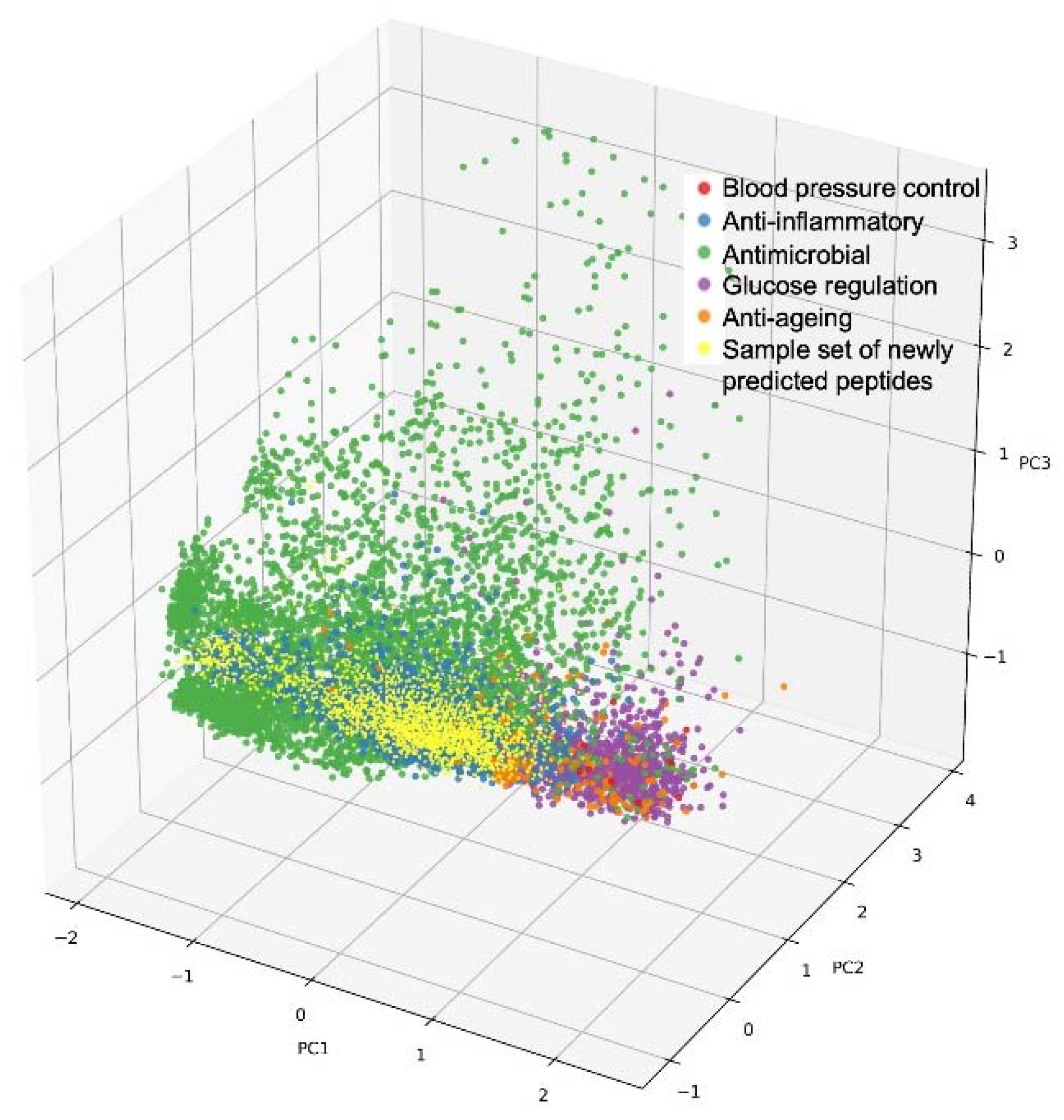
Figure 1. Illustration of a multi-dimensional peptide property space: 3D projection of annotated bioactive peptides according to three principal components (PC1-3), visualized as colored dots according to different health benefits. Red: blood pressure control; blue: anti-inflammatory; green: anti-microbial; magenta: glucose regulation; orange: anti-ageing; yellow: sample set of newly predicted peptides.
The above-described peptide characteristics encompass the biological, biochemical, and physical properties, but they can also include peptide suitability and applicability resulting from these primary properties [24][15]. The biological, biochemical, and physical properties encompass amino acid sequence, molecular weight, hydrophobicity/hydrophilicity, basicity/acidity, solubility, and biochemical stability [36][24]. These features can determine the peptide’s suitability for specific applications. The key qualities for orally administered bioactive peptides are, for example, resistance to gastrointestinal digestion; transportability from the gut lumen to systemic blood circulation (active or passive transport across biological membranes); or the stability and half-life in human blood once absorbed from gut to blood [5][25]. The primary peptide properties may also determine the peptide’s relevance for commercial translation: the uniqueness of the peptide sequence influences the protectability of intellectual property; the amino acid sequence and length of a peptide impacts the manufacturing costs [37][26].
2.3. Natural Peptide Network (NPN) Design and Validation of Predicted Peptides and Designed Hydrolysates
Once a feasible and potent set of in vitro bioactive peptides has been identified and consolidated (from ‘hits to leads’), these candidate peptides can be blasted against all known plant and food genomes, proteomes, and peptidomes [36][24]. The purpose of this straightforward exercise is to identify the best natural source that holds the majority, if not all, of the desired peptide sequences in the plant and food parent proteins. When this optimal source is found—be it a grain, legume, vegetable, or any other edible plant, including not only terrestrial but also marine sources—the best protein hydrolysate with the resulting Natural Peptide Network (NPN), i.e., the combination of the predicted and initially validated peptide leads, needs to be designed [37][26]. Proteolytic digestion as such, be it in vivo (digestion) or in vitro (processing), may but does not necessarily produce bioactive peptides because these processes are not designed to do so. However, once a set of bioactive peptides deriving from plant and food proteins has been identified, the in vitro hydrolysis can be optimized to release the desired bioactive peptides, in addition to many others. This process represents a standard bioinformatic sequence analysis in the sense that the peptide locations in their parent proteins are determined and the adjacent potential cleavage sites are identified. With the latter in hand, the best proteases can be identified to be applied in combination to result in an optimized hydrolysis process and eventually yield the right combination and concentration of the targeted bioactive peptides [36][24]. In short, the best plant/food source in terms of the presence and abundance of the lead peptides in parent proteins is identified; based on the lead peptide sequence and its position within the parent protein in the plant/food source, the best enzymes for specific release of the lead peptides from their parent proteins are selected.
The predicted peptides with the best initial in vitro performance are then tested more extensively in relevant in vitro assays for bioactivity and toxicity. These peptides should be tested both individually and as the blend generated in the protein hydrolysate. Often, the hydrolysate exhibits greater bioactivity than the individual peptides, thanks to the combined, or even additive, effects of the multiple active principles present in the hydrolysate. These tests are typically performed over a range of peptide and hydrolysate concentrations, which can already to some extent inform on subsequent in vivo and in human studies. The advantage of bioactive food peptides is their inherent biosafety [8][27]. Therefore, after successful in vitro validation, the peptide and/or hydrolysate testing can often directly advance to a pilot pre-clinical study.
2.4. Natural Peptide Network (NPN) Manufacturing and Analysis
The optimal plant/food source is insourced, and the bulk protein material is extracted. The identified enzymes (proteases) are applied to a solution/suspension of this bulk protein material in different combinations and under different conditions (temperature, pH, time, concentration, and so etc.on) [30,31][18][19]. The resulting protein hydrolysates are analyzed by MS-based peptidomics, which delivers a detailed peptide profile encompassing the NPN, i.e., the nearly complete hydrolysate peptidome [38][28]. Mass spectrometry enables the sequencing, identifying, and quantifying of peptides and proteins and is described in detail in the next section. Based on these preparations and analyses, the best procedure for manufacturing the desired protein hydrolysate with the optimal peptide profile is identified. The reproducibility of this optimized procedure is established in replicate manufacturing and analysis experiments. The optimized procedure is then established and used for further laboratory scale preparations, as well as for the pilot- and large-scale hydrolysate production for clinical trials and commercial purposes, respectively [32][20].
References
- Conway, A.R.A.; Kovacs, K. New and emerging models of human intelligence. Wiley Interdiscip. Rev. Cogn. Sci. 2015, 6, 419–426.
- Coen, E. The storytelling arms race: Origin of human intelligence and the scientific mind. Heredity 2019, 123, 67–78.
- Dzobo, K.; Adotey, S.; Thomford, N.E.; Dzobo, W. Integrating Artificial and Human Intelligence: A Partnership for Responsible Innovation in Biomedical Engineering and Medicine. OMICS 2020, 24, 247–263.
- Abdelhalim, H.; Berber, A.; Lodi, M.; Jain, R.; Nair, A.; Pappu, A.; Patel, K.; Venkat, V.; Venkatesan, C.; Wable, R.; et al. Artificial Intelligence, Healthcare, Clinical Genomics, and Pharmacogenomics Approaches in Precision Medicine. Front. Genet. 2022, 13, 929736.
- Limketkai, B.N.; Mauldin, K.; Manitius, N.; Jalilian, L.; Salonen, B.R. The Age of Artificial Intelligence: Use of Digital Technology in Clinical Nutrition. Curr. Surg. Rep. 2021, 9, 20.
- Walter, W.; Haferlach, C.; Nadarajah, N.; Schmidts, I.; Kühn, C.; Kern, W.; Haferlach, T. How artificial intelligence might disrupt diagnostics in hematology in the near future. Oncogene 2021, 40, 4271–4280.
- Harfouche, A.L.J.; Jacobson, D.A.; Kainer, D.; Romero, J.C.; Harfouche, A.H.; Mugnozza, G.S.; Moshelion, M.; Tuskan, G.A.; Keurentjes, J.J.B.; Altman, A. Accelerating Climate Resilient Plant Breeding by Applying Next-Generation Artificial Intelligence. Trends Biotechnol. 2019, 37, 1217–1235.
- Zhao, S.; Xie, C. Spatial Difference of China’s Regional Logistics Development and Construction of Information Network Platform Based on Artificial Intelligence Technology Under the Background of New Economy. Front. Psychol. 2022, 13, 871538.
- Sarker, S.; Jamal, L.; Ahmed, S.F.; Irtisam, N. Robotics and artificial intelligence in healthcare during COVID-19 pandemic: A systematic review. Robot. Auton. Syst. 2021, 146, 103902.
- Schneider, P.; Walters, W.P.; Plowright, A.T.; Sieroka, N.; Listgarten, J.; Goodnow, R.A., Jr.; Fisher, J.; Jansen, J.M.; Duca, J.S.; Rush, T.S.; et al. Rethinking drug design in the artificial intelligence era. Nat. Rev. Drug Discov. 2019, 19, 353–364.
- Nam, D.; Chapiro, J.; Paradis, V.; Seraphin, T.P.; Kather, J.N. Artificial intelligence in liver diseases: Improving diagnostics, prognostics and response prediction. JHEP Rep. 2022, 4, 100443.
- Luccioni, A.; Schmidt, V.; Vardanyan, V.; Bengio, Y.; Rhyne, T.-M. Using Artificial Intelligence to Visualize the Impacts of Climate Change. IEEE Comput. Graph. Appl. 2021, 41, 8–14.
- Doherty, A.; Wall, A.; Khaldi, N. Using Artificial Intelligence to Reduce Global Healthcare Costs through Discovery and Development of Nutritional Interventions. Int. J. Nurs. Didact. 2020, 10, 1–5.
- Doherty, A.; Wall, A.; Khaldi, N.; Kussmann, M. Artificial Intelligence in Functional Food Ingredient Discovery and Characterisation: A Focus on Bioactive Plant and Food Peptides. Front. Genet. 2021, 12, 768979.
- Minkiewicz, P.; Dziuba, J.; Iwaniak, A.; Dziuba, M.; Darewicz, M. BIOPEP database and other programs for processing bioactive peptide sequences. J. AOAC Int. 2008, 91, 965–980.
- Zhang, Y.; Li, P.; Ma, Y.; Wang, J.; Chen, Y. Artificial intelligence accelerates the mining of bioactive small molecules from human microbiome. Clin. Transl. Med. 2022, 12, e1011.
- Casey, R.; Adelfio, A.; Connolly, M.; Wall, A.; Holyer, I.; Khaldi, N. Discovery through Machine Learning and Preclinical Validation of Novel Anti-Diabetic Peptides. Biomedicines 2021, 9, 276.
- Corrochano, A.R.; Cal, R.; Kennedy, K.; Wall, A.; Murphy, N.; Trajkovic, S.; O’Callaghan, S.; Adelfio, A.; Khaldi, N. Characterising the efficacy and bioavailability of bioactive peptides identified for attenuating muscle atrophy within a Vicia faba-derived functional ingredient. Curr. Res. Food Sci. 2021, 4, 224–232.
- Kennedy, K.; Keogh, B.; Lopez, C.; Adelfio, A.; Molloy, B.; Kerr, A.; Wall, A.M.; Jalowicki, G.; Holton, T.A.; Khaldi, N. An Artificial Intelligence Characterised Functional Ingredient, Derived from Rice, Inhibits TNF-α and Significantly Improves Physical Strength in an Inflammaging Population. Foods 2020, 9, 1147.
- Chauhan, S.; Kerr, A.; Keogh, B.; Nolan, S.; Casey, R.; Adelfio, A.; Murphy, N.; Doherty, A.; Davis, H.; Wall, A.; et al. An Artificial-Intelligence-Discovered Functional Ingredient, NRT_N0G5IJ, Derived from Pisum sativum, Decreases HbA1c in a Prediabetic Population. Nutrients 2021, 13, 1635.
- van Erp, M.; Reynolds, C.; Maynard, D.; Starke, A.; Martín, R.I.; Andres, F.; Leite, M.C.A.; de Toledo, D.A.; Rivera, X.S.; Trattner, C.; et al. Using Natural Language Processing and Artificial Intelligence to Explore the Nutrition and Sustainability of Recipes and Food. Front. Artif. Intell. 2020, 3, 621577.
- Sánchez-Ruiz, A.; Colmenarejo, G. Systematic Analysis and Prediction of the Target Space of Bioactive Food Compounds: Filling the Chemobiological Gaps. J. Chem. Inf. Model. 2022, 62, 3734–3751.
- Li, G.; Shuli, Z.; Linlin, L.; Yinghu, Z.; Nan, S.; Haibin, W.; Hongyu, X. Bioinformatics and Computer Simulation approaches to the discovery and analysis of Bioactive Peptides. Curr. Pharm. Biotechnol. 2022, 23, 1541–1555.
- Minkiewicz, P.; Darewicz, M.; Iwaniak, A.; Sokołowska, J.; Starowicz, P.; Bucholska, J.; Hrynkiewicz, M. Common Amino Acid Subsequences in a Universal Proteome—Relevance for Food Science. Int. J. Mol. Sci. 2015, 16, 20748–20773.
- Karami, Z.; Akbari-Adergani, B. Bioactive food derived peptides: A review on correlation between structure of bioactive peptides and their functional properties. J. Food Sci. Technol. 2019, 56, 535–547.
- Duffuler, P.; Bhullar, K.S.; de Campos Zani, S.C.; Wu, J. Bioactive Peptides: From Basic Research to Clinical Trials and Commercialization. J. Agric. Food Chem. 2022, 70, 3585–3595.
- Schaafsma, G. Safety of protein hydrolysates, fractions thereof and bioactive peptides in human nutrition. Eur. J. Clin. Nutr. 2009, 63, 1161–1168.
- Fan, K.-T.; Hsu, C.-W.; Chen, Y.-R. Mass spectrometry in the discovery of peptides involved in intercellular communication: From targeted to untargeted peptidomics approaches. Mass Spectrom. Rev. 2022, 41, e21789.
More
