Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Amina Yu and Version 1 by Chun-Hui Gao.
In nature, DNA is ubiquitous, existing not only inside but also outside of the cells of organisms. Intracellular DNA (iDNA) plays an essential role in different stages of biological growth, and it is defined as the carrier of genetic information. In addition, extracellular DNA (eDNA) is not enclosed in living cells, accounting for a large proportion of total DNA in the environment. Both the lysis-dependent and lysis-independent pathways are involved in eDNA release, and the released DNA has diverse environmental functions.
- extracellular DNA
- origin
- functions
- distribution
- organisms
1. Introduction
A general consensus has risen that the total DNA in a natural environment is comprised of intracellular DNA (iDNA) and extracellular DNA (eDNA). DNA has been thought to exist only inside the cell for long periods of time. However, an increasing amount of evidence has shown the presence of DNA in the extracellular space, and these eDNA outside the cell may account for a large proportion of the total DNA [1]. Compared with iDNA that is located in living cells, eDNA is outside the cell, and it is widely distributed in soil, sediments, feces and aquatic ecosystems and other ecological niches, and it has also been discovered in tissue cultures and in the blood of the human body and some other animals [2,3,4,5][2][3][4][5]. Moreover, the ratio of eDNA:iDNA could provide valuable information about the microbial activity in different environments [6[6][7],7], although the activity of different microorganisms is not completely correlated with the ratio [8]. These results indicate that eDNA has the potential to be a proxy for specific microbial activities [9]. Initially, eDNA was thought to be derived from lysed cells. Nowadays, it has been well documented that both the lysis-dependent and lysis-independent pathways were involved in eDNA release in prokaryotic and eukaryotic cells [10]. There are two general forms of eDNA, which are single-stranded DNA (ssDNA) and double-stranded DNA (dsDNA), and the latter accounts for a significant portion of the total eDNA in the environment due to the instability of ssDNA [11]. The increasing amount of evidence has shown that eDNA plays a significant role in multiple biological processes such as biofilm formation and maturation, horizontal gene transfer (HGT), and multiple element cycling including for carbon, nitrogen, and phosphate [12,13,14,15][12][13][14][15]. However, the biological roles of eDNA are different across different species, and they even vary with the different life stages in the same organism. For example, in Neisseria meningitidis, eDNA has different roles in early and late biofilm formation [16]. During the past decades, biodiversity is disappearing at an alarming rate, and the large-scale assessment and monitoring of biodiversity has become a priority. Therefore, eDNA-based biomonitoring has emerged as an effective tool for detecting biodiversity changes at a large scale, and it offers us a better understanding of the microbial community composition as well as the community function [9].eDNA is widely distributed in the soil, sediments, feces, and other environments (Figure 31).
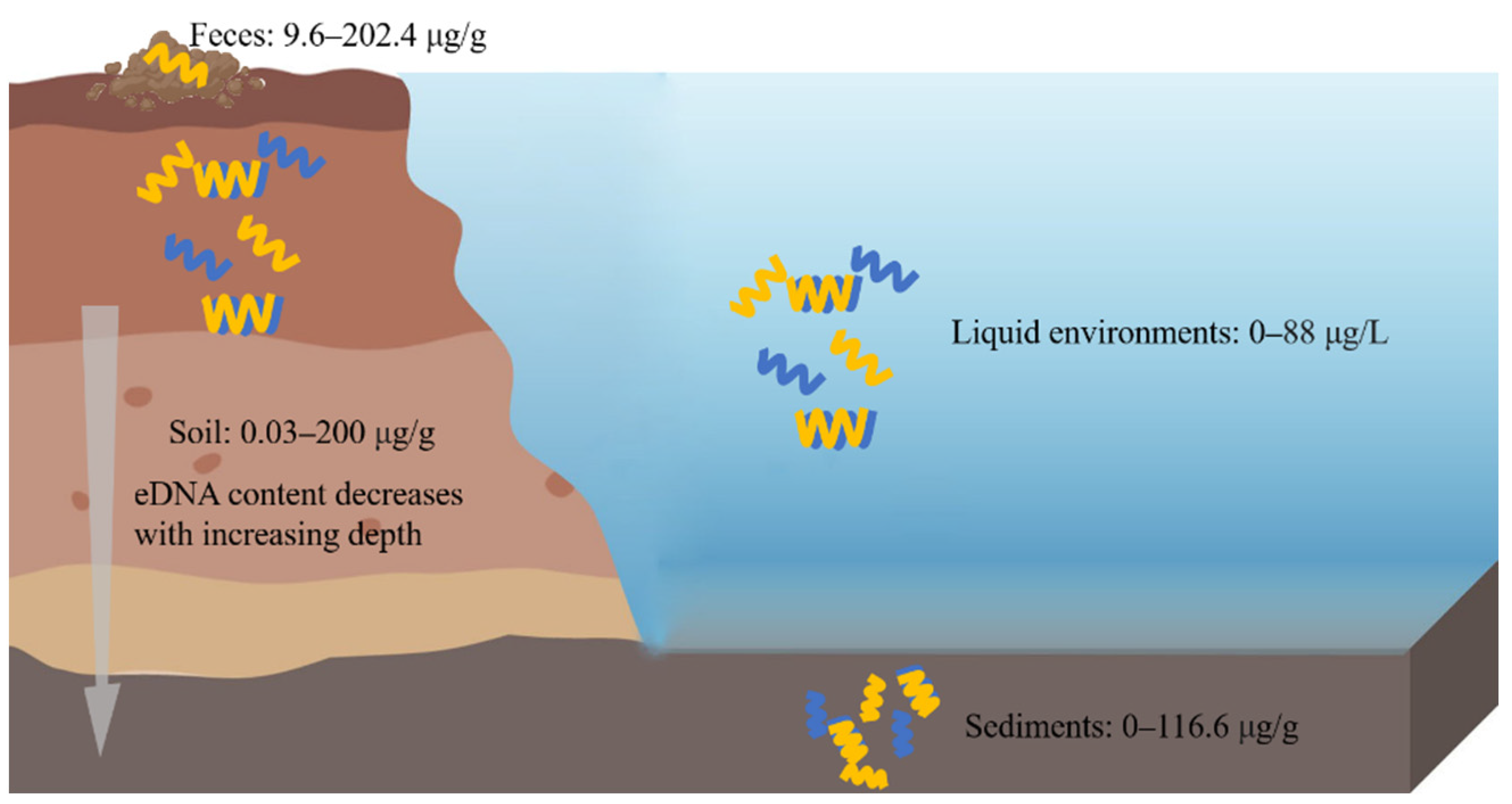
Figure 31. eDNA in different ecological environments. eDNA is widely distributed in soil, sediments, feces, and liquid environments. The left side of the picture represents the soil, and above the soil are the feces, and on the right side of the picture are the liquid environments, and under these is the sediment.
2. eDNA in Soil
eDNA widely exists in soil, and it accounts for about 40% of the total DNA pool. Its content of eDNA varies significantly in different soil regions, types, and layers, and most of the eDNA is concentrated in the upper layers of the soil [15]. The range of eDNA content in soil is about 0.03–200 μg/g [53][17]. At the increasing depth of the soil layer, the eDNA content tends to decrease, and its reduction is much more obvious than that of organic carbon [1]. In addition, it has been found that the higher the fertilizer supply is, then the lower the eDNA concentration is [62][18]. It is estimated that more than 70% of the DNA molecules present in soil are derived from fungi [63][19].
The eDNA in soil can be divided into two fractions with one that is tightly bound to the soil particles and the other that is weakly bound [64][20] so that the eDNA can be protected against degradation by nucleases in the soil, thus, stably persisting in the soil for a long time [53][17]. Therefore, eDNA can serve as a historical microbial gene reservoir, reflecting the diachronic biodiversity of the investigated environments. The long-term stable existence of eDNA in soil can lead to the accumulation of exogenous genes such as antibiotic resistance genes (ARGs) which may be passed between cells, meanwhile soil microorganisms acquire ARGs through homologous recombination, thus, potentially generating new resistance strains, especially resistant pathogenic bacteria. These strains have strong resistance and are not easily subjected to environmental stresses, which pose great threats to the environments [65][21]. In addition, eDNA occupies a large proportion (about 10%) of the soil P pool, which enables the microbes to take up DNA as a nutrition source of carbon, nitrogen, phosphorus, and nucleic acid precursors [66][22]. Surprisingly, adding eDNA to a culture medium has been reported to promote the growth of the lateral roots and root hairs in Arabidopsis [67][23]. However, it has been found that conspecific eDNA treatment can inhibit the growth of plants and the soil microbe, whereas heterologous eDNA treatment cannot, indicating that this inhibitory effect might be due to the maintenance of the microbial diversity [68][24]. However, more research is needed to further explore the underlying mechanism. In addition, plants can release NETs in which eDNA is able to enhance the resistance of root tip to soil pathogens, but the resistance of these pathogens would be lost with a DNaseI treatment [69][25].
3. eDNA in Sediments
eDNA can be found in various types of sediments, such as marine sediments, river sediments and freshwater sediments. Among them, the marine sediments are the largest sediment eDNA reservoir. Increasing evidence suggests that more than 90% of DNA in sediments is extracellular, and it has been estimated that the content of eDNA ranges from 0.30 to 0.45 Gt in deep-sea sediments [70][26]. The content of eDNA varies in the different sediments. A study of Haihe River sediments has demonstrated that the concentration of eDNA (96.8 ± 19.8 μg/g) is much higher that of iDNA (76.7 ± 13.0 μg/g) [71][27]. In ferruginous sediments from Lake Towuti of Indonesia, the eDNA concentration is at around 0.5–0.6 μg/g in the surface layer of the sediments, and the concentration of eDNA has the same trends in the sediments and soil, namely, it decreases with the increasing depth [72][28]. Apart from the eDNA concentration, the eDNA fragment size also decreases as the depth increases [5].
The most of the eDNA in the sediments is bound or adsorbed to particles to fight against nuclease degradation [73][29]. Additionally, the eDNA in sediments possesses multiple roles. For example, the eDNA in the marine sediments can not only reflect the biogenesis processes on different time scales, but also provide information on biodiversity and genetic diversity of different ecosystems in both ancient and modern times [74][30]. In addition, eDNA could be used to obtain past climate and environmental information through a taxa analysis, and it promises to be a very powerful tool for predicting future environmental changes and the function of the ecosystem [75][31].
4. eDNA in Feces
eDNA can be extracted from cattle feces, and its content is about 202.4 μg/g from fresh excrements [76][32]. Additionally, the content of eDNA in swine mature ranges from 9.6 to 9.7 μg/g dw (dry weight) [77][33]. There findings indicate that eDNA is an essential ARGs reservoir. eDNA receives increasing attention since the ARG pool in mature animals poses a threat to the ecological environments and human health. Livestock and poultry manure is generally considered to be a natural host for ARGs, and the environment exposure of manure from farms may drive the circulation of ARGs [78][34]. In humans, gut microbiome dysregulation may lead to the occurrence of a series of diseases, and thus, extracellular viral-like particles (eVLPs) and virus DNA from human feces could be extracted for metagenome and morphology analyses [79,80][35][36].
5. eDNA in Other Ecological Environments
eDNA exists in various liquid environments such as seawater, freshwater, river water, and lake water. The eDNA concentration in aquatic environments can reach up to 88 μg/L [81][37]. The concentration of eDNA is always associated with the trophic status and the season with its concentrations ranging from 2.5 to 46 μg/L in mesotrophic water, while it ranged from 11.5 to 72 μg/L in eutrophic water [82][38].
References
- Bairoliya, S.; Xiang, J.K.Z.; Cao, B. Extracellular DNA in Environmental Samples: Occurrence, Extraction, Quantification, and Impact on Microbial Biodiversity Assessment. Appl. Environ. Microbiol. 2022, 88, e01845-21.
- Fuchs, T.A.; Brill, A.; Duerschmied, D.; Schatzberg, D.; Monestier, M.; Myers, D.D.; Wrobleski, S.K.; Wakefield, T.W.; Hartwig, J.H.; Wagner, D.D. Extracellular DNA traps promote thrombosis. Proc. Natl. Acad. Sci. USA 2010, 107, 15880–15885.
- Gartler, S.M. Cellular uptake of deoxyribonucleic acid by human tissue culture cells. Nature 1959, 184 (Suppl. S19), 1505–1506.
- Mandel, P.; Metais, P.; Métais, P. Les acides nucléiques du plasma sanguin chez l’homme. Cr. Acad. Sci. Paris 1948, 142, 241–243.
- Torti, A.; Jørgensen, B.B.; Lever, M.A. Preservation of microbial DNA in marine sediments: Insights from extracellular DNA pools. Environ. Microbiol. 2018, 20, 4526–4542.
- Gómez-Brandón, M.; Ascher-Jenull, J.; Bardelli, T.; Fornasier, F.; Sartori, G.; Pietramellara, G.; Arfaioli, P.; Egli, M.; Beylich, A.; Insam, H.; et al. Ground cover and slope exposure effects on micro- and mesobiota in forest soils. Ecol. Indic. 2017, 80, 174–185.
- Gómez-Brandón, M.; Ascher-Jenull, J.; Bardelli, T.; Fornasier, F.; Fravolini, G.; Arfaioli, P.; Ceccherini, M.T.; Pietramellara, G.; Lamorski, K.; Sławiński, C.; et al. Physico-chemical and microbiological evidence of exposure effects on Picea abies—Coarse woody debris at different stages of decay. Forest Ecol. Manag. 2017, 391, 376–389.
- Nagler, M.; Podmirseg, S.M.; Griffith, G.W.; Insam, H.; Ascher-Jenull, J. The use of extracellular DNA as a proxy for specific microbial activity. Appl. Microbiol. Biot. 2018, 102, 2885–2898.
- Nagler, M.; Insam, H.; Pietramellara, G.; Ascher-Jenull, J. Extracellular DNA in natural environments: Features, relevance and applications. Appl. Microbiol. Biot. 2018, 102, 6343–6356.
- Campoccia, D.; Montanaro, L.; Arciola, C.R. Tracing the origins of extracellular DNA in bacterial biofilms: Story of death and predation to community benefit. Biofouling 2021, 37, 1022–1039.
- Pallares, R.M.; Thanh, N.T.K.; Su, X. Tunable plasmonic colorimetric assay with inverse. sensitivity for extracellular DNA quantification. Chem. Commun. 2018, 54, 11260–11263.
- Dominiak, D.M.; Nielsen, J.L.; Nielsen, P.H. Extracellular DNA is abundant and important for microcolony strength in mixed microbial biofilms. Environ. Microbiol. 2011, 13, 710–721.
- Flemming, H.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633.
- Mulcahy, H.; Charron-Mazenod, L.; Lewenza, S. Pseudomonas aeruginosa produces an extracellular deoxyribonuclease that is required for utilization of DNA as a nutrient source. Environ. Microbiol. 2010, 12, 1621–1629.
- Nielsen, K.M.; Johnsen, P.J.; Bensasson, D.; Daffonchio, D. Release and persistence of extracellular DNA in the environment. Environ. Biosaf. Res. 2007, 6, 37–53.
- Lappann, M.; Claus, H.; van Alen, T.; Harmsen, M.; Elias, J.; Molin, S.; Vogel, U. A dual role of extracellular DNA during biofilm formation of Neisseria meningitidis. Mol. Microbiol. 2010, 75, 1355–1371.
- Pietramellara, G.; Ascher, J.; Borgogni, F.; Ceccherini, M.T.; Guerri, G.; Nannipieri, P. Extracellular DNA in soil and sediment: Fate and ecological relevance. Biol. Fert. Soils 2009, 45, 219–235.
- Niemeyer, J.; Gessler, F. Determination of free DNA in soils. J. Plant Nutr. Soil Sc. 2002, 165, 121–124.
- Borneman, J.; Hartin, R.J. PCR Primers That Amplify Fungal rRNA Genes from Environmental Samples. Appl. Environ. Microb. 2000, 66, 4356–4360.
- Pathan, S.I.; Arfaioli, P.; Ceccherini, M.T.; Ascher-Jenull, J.; Pietramellara, G. Preliminary evidences of the presence of extracellular DNA single stranded forms in soil. PLoS ONE 2020, 15, e0227296.
- Poté, J.; Ceccherini, M.T.; Van, V.T.; Rosselli, W.; Wildi, W.; Simonet, P.; Vogel, T.M. Fate and transport of antibiotic resistance genes in saturated soil columns. Eur. J. Soil Biol. 2003, 39, 65–71.
- Levy-Booth, D.J.; Campbell, R.G.; Gulden, R.H.; Hart, M.M.; Powell, J.R.; Klironomos, J.N.; Pauls, K.P.; Swanton, C.J.; Trevors, J.T.; Dunfield, K.E. Cycling of extracellular DNA in the soil environment. Soil Biol. Biochem. 2007, 39, 2977–2991.
- Paungfoo-Lonhienne, C.; Lonhienne, T.G.A.; Schmidt, S. DNA uptake by Arabidopsis induces changes in the expression of CLE peptides which control root morphology. Plant Signal. Behav. 2010, 153, 799–805.
- Mazzoleni, S.; Bonanomi, G.; Incerti, G.; Chiusano, M.L.; Termolino, P.; Mingo, A.; Senatore, M.; Giannino, F.; Cartenì, F.; Rietkerk, M.; et al. Inhibitory and toxic effects of extracellular self-DNA in litter: A mechanism for negative plant–soil feedbacks? New Phytol. 2015, 205, 1195–1210.
- Hawes, M.; McLain, J.; Ramirez-Andreotta, M.; Curlango-Rivera, G.; Flores-Lara, Y.; Brigham, L. Extracellular Trapping of Soil Contaminants by Root Border Cells: New Insights into Plant Defense. Agronomy 2016, 6, 5.
- Dell’Anno, A.; Danovaro, R. Extracellular DNA Plays a Key Role in Deep-Sea Ecosystem Functioning. Science 2005, 309, 2179.
- Mao, D.; Luo, Y.; Mathieu, J.; Wang, Q.; Feng, L.; Mu, Q.; Feng, C.; Alvarez, P.J.J. Persistence of Extracellular DNA in River Sediment Facilitates Antibiotic Resistance Gene Propagation. Environ. Sci. Technol. 2014, 48, 71–78.
- Vuillemin, A.; Horn, F.; Alawi, M.; Henny, C.; Wagner, D.; Crowe, S.A.; Kallmeyer, J. Preservation and Significance of Extracellular DNA in Ferruginous Sediments from Lake Towuti, Indonesia. Front. Microbiol. 2017, 8, 1440.
- Dell’Anno, A.; Bompadre, S.; Danovaro, R. Quantification, base composition, and fate of extracellular DNA in marine sediments. Limnol. Oceanogr. 2002, 47, 899–905.
- Dell’Anno, A.; Corinaldesi, C.; Stavrakakis, S.; Lykousis, V.; Danovaro, R. Pelagic-Benthic Coupling and Diagenesis of Nucleic Acids in a Deep-Sea Continental Margin and an Open-Slope System of the Eastern Mediterranean. Appl. Environ. Microb. 2005, 71, 6070–6076.
- Ellegaard, M.; Clokie, M.R.J.; Czypionka, T.; Frisch, D.; Godhe, A.; Kremp, A.; Letarov, A.; McGenity, T.J.; Ribeiro, S.; John Anderson, N. Dead or alive: Sediment DNA archives as tools for tracking aquatic evolution and adaptation. Commun. Biol. 2020, 3, 169.
- Chroňáková, A.; Ascher, J.; Jirout, J.; Ceccherini, M.T.; Elhottová, D.; Pietramellara, G.; Šimek, M. Cattle impact on composition of archaeal, bacterial, and fungal communities by comparative fingerprinting of total and extracellular DNA. Biol. Fert. Soils 2013, 49, 351–361.
- Dong, P.; Wang, H.; Fang, T.; Wang, Y.; Ye, Q. Assessment of extracellular antibiotic resistance genes (eARGs) in typical environmental samples and the transforming ability of eARG. Environ. Int. 2019, 125, 90–96.
- Xu, Y.; Li, H.; Shi, R.; Lv, J.; Li, B.; Yang, F.; Zheng, X.; Xu, J. Antibiotic resistance genes in different animal manures and their derived organic fertilizer. Environ. Sci. Eur. 2020, 32, 102.
- Castro-Mejia, J.L.; Deng, L.; Vogensen, F.K.; Reyes, A.; Nielsen, D.S. Extraction and Purification of Viruses from Fecal Samples for Metagenome and Morphology Analyses. Methods Mol. Biol. 2018, 1838, 49–57.
- Wu, H.; Tremaroli, V.; Bäckhed, F. Linking Microbiota to Human Diseases: A Systems Biology Perspective. Trends Endocrinol. Metab. 2015, 26, 758–770.
- Liang, Z.; Keeley, A. Filtration Recovery of Extracellular DNA from Environmental Water Samples. Environ. Sci. Technol. 2013, 47, 9324–9331.
- Waldemar Siuda, H.G. Determination of dissolved deoxyribonucleic acid concentration in lake water. Aquat. Microb. Ecol. 1996, 11, 193–202.
More
