Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Conner Chen and Version 1 by Irene Dini.
Antimicrobial peptides are made by lower and higher organisms responding to pathogenic challenges. [12].Antimicrobial peptides (AMPs) kill the invading pathogens and modulate the innate immune response. They are commonly classified according to their sources, amino-acid-rich species, structural characteristics, and activities [13]. In multicellular organisms and humans, they are localized into specific sites commonly exposed to microbes (i.e., mucosa epithelia and skin).
- AMPs
- food preservation
- food shelf-life
- active packaging
- Gram-positive bacteria
1. Viral AMPs
Some phage proteins, including lysins, depolymerases, virion-associated peptidoglycan hydrolases (VAPGHs), and holins, show antibacterial activity [14][1]. They are defined as “enzybiotics” to indicate their use as antibacterial materials as alternatives to standard antibiotics [15][2]. The two types of phage Antimicrobial peptides (AMPs) are known as phage-encoded lytic factors and phage-tail complexes [16][3].
Phage lysines (size range from 25 to 40 kDa) are peptidoglycan-hydrolyzing enzymes [17][4], which can hydrolyze the microbial cell wall, permitting bacteriophage progeny release [16][3]. Lysins have rapid bactericidal activity (against Gram-positive and Gram-negative bacteria) and other desirable characteristics, such as synergy with cell-wall-reducing antibiotics, anti-biofilm action, heat stability up to ~50 °C, and the possibility of lyophilization [18,19,20][5][6][7]. Peptidoglycan hydrolases (VAPGHs), encoded mainly by double-stranded DNA phages, have high thermal stability. They infect Gram-positive and Gram-negative bacteria. VAPGHs have a C-terminal cell-wall-binding domain, which can link them to receptors on the bacterial cell surface. They inject genetic materials into bacterial cells after partially and locally damaging bacterial cell wall peptidoglycans [21][8]. They can be classified into three categories: glycosidases that cut glycosidic bonds in the peptidoglycan chain, amidases that cut amide bonds (between N-acetylmuramic acid lactyl and stem peptide l-alanines), and endopeptidases that cleave peptide bonds within either the stem peptide or cross-link [22][9].
Phage-tail-like AMPs are high-molecular-weight cylindrical peptides with a structure like a phage tail [23,24][10][11]. They can be classified into two classes: R-type (related to Myoviridae phage tails) and F-type (related to Siphoviridae phage tails) [23][10].
R-type phage-tail-like bacteriocins are nonflexible and have tubes surrounded by contractile sheaths [25][12]. They initially make a channel in the cell membrane and successively drive their internal core into the cell. This process determines rapid cell death by decoupling cellular ion gradients [23][10], interfering with oxygen uptake, and affecting macromolecule synthesis [26][13].
F-type phage-tail-like bacteriocins are flexible and noncontractile [25][12]. They act similarly to R-type bacteriocins [27][14].
2. Bacterial AMPs
2.1. AMPs Made by Gram-Positive Bacteria
Gram-positive bacteria can produce AMPs in ribosomes (ribosomal AMPs) or enzymatically (non-ribosomal AMPs) [28,29][15][16] (Figure 21). Twenty-sixty amino acids (hydrophobic and cationic) can make up ribosomally synthesized bacterial AMPs (bacteriocins) [30][17].
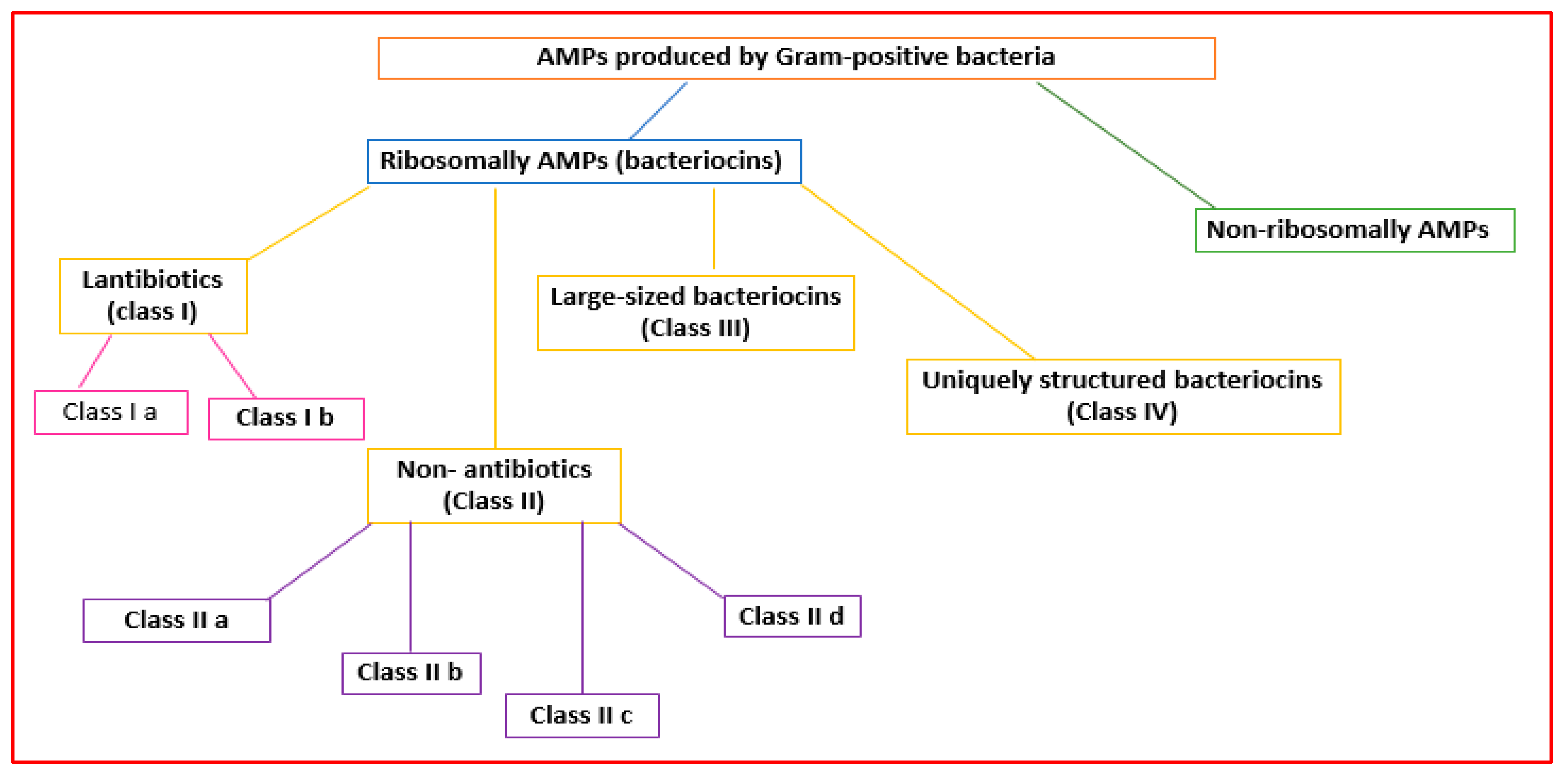
Figure 21.
Classification of the AMPs produced by Gram-positive bacteria.
Bacteriocins can be classified into bacteriocins produced by Gram-positive and Gram-negative bacteria [31][18].
2.2. AMPs Made by Gram-Positive Bacteria
Gram-positive organisms make bacteriocins that can be grouped into lantibiotics (class I), non-lantibiotics (class II), large-sized bacteriocins (class III), and uniquely structured bacteriocins (class IV) [32][19] (Figure 21).
Lantibiotics are active against primarily Gram-positive bacteria [32][19]. They are small peptides (<5 kDa; 19–50 amino acids) that are stable to heat, pH, and proteolysis [33][20]. Lantibiotics can be subdivided into subclasses Ia and Ib (Figure 21).
Subclass Ib lantibiotics are inflexible peptides that decrease the activity of bacteria crucial enzymes [32][19].
Class II AMPs (non-lanthionine-containing bacteriocins) are small (<10 kDa) and heat-stable peptides that can form pores in the bacterial membrane. They can be grouped into four subclasses [35][22].
Subclass IIa consists of disulfide linear peptides with similar amino acid sequences that permeabilize the cell membrane, showing significant antilisterial activity [36][23].
Subclass IIb bacteriocins increase the permeability of the bacterial cell membrane to specific small molecules [37][24]. They contain two peptide subunits (α and β) [37][24].
Subclass IIc bacteriocins permeabilize the microbial membrane, dissipate the membrane potential, and cause cell death [38][25]. They comprise small, cyclic peptides whose C- and N-terminals are covalently linked [39][26].
Class III bacteriocins (bacteriolysins) [35][22] are large (>30 kDa), heat-labile peptides [32][19] (Figure 21).
Class IV AMPs containing lipids or carbohydrates are susceptible to lipolytic and glycolytic enzymes [40][27] (Figure 21).
Non-ribosomally synthesized AMPs are made from peptide synthetases produced by Gram-positive and Gram-negative bacteria [9][28].
2.3. AMPs Made by Gram-Negative Bacteria
Gram-negative organisms make bacteriocins that can be grouped into microcins, colicins, colicin-like bacteriocins, and phage-tail-like bacteriocins [41][29] (Figure 32).
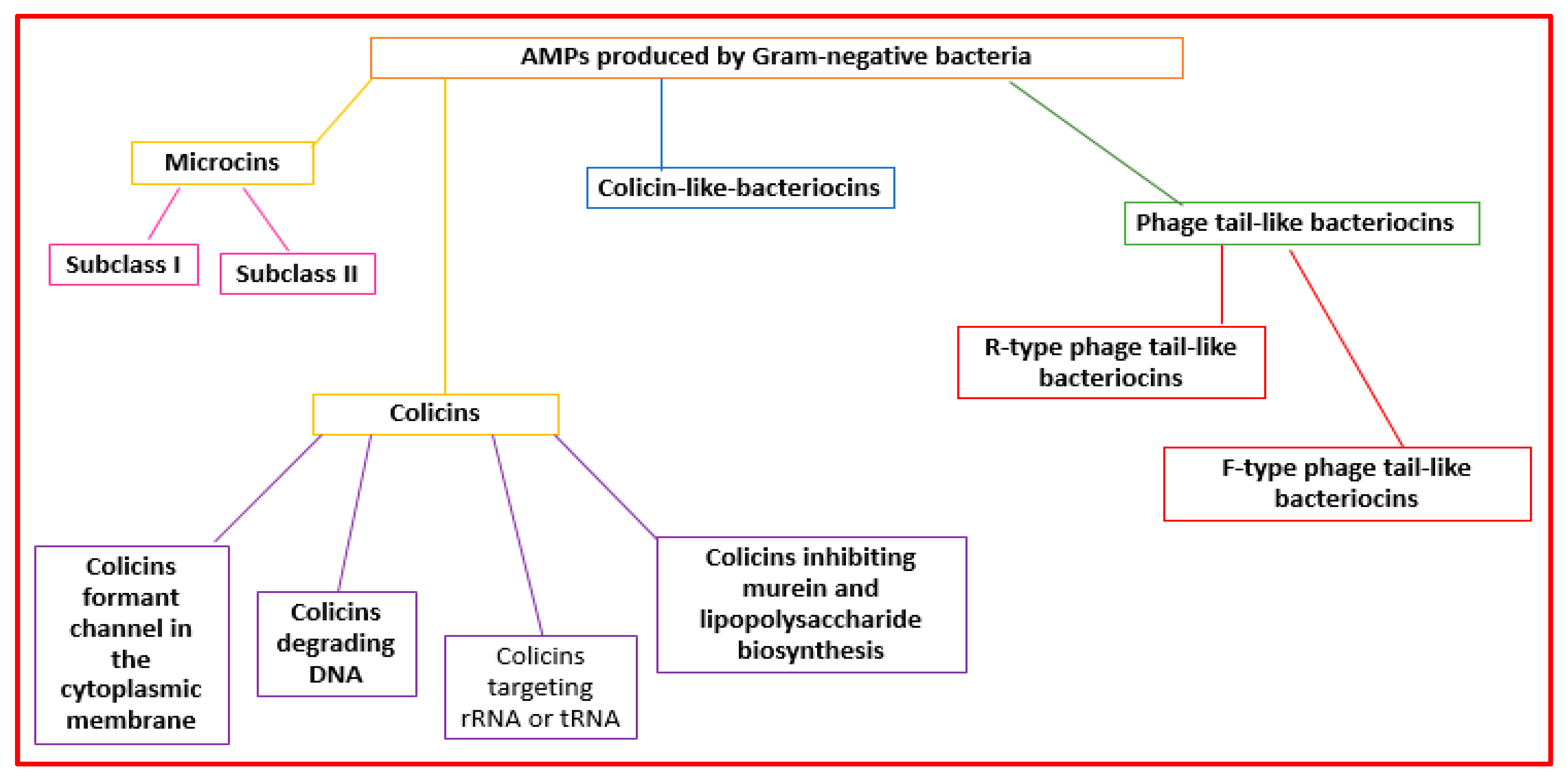
Figure 32.
Classification of the AMPs produced by Gram-negative bacteria.
Microcins are made by Enterobacteriaceae. Microcins interact with some cellular targets. They can format pores that determine membrane disruption [23][10] or decrease the functionality of enzymes (the ATP synthase complex, DNA gyrase, RNA polymerase, and aspartyl-t RNA synthetase) [32][19]. They are grouped into two subclasses: subclass I (molecular weight lower than 5 kDa) and subclass II (molecular weight ranging from 5 to 10 kDa) [42,43][30][31] (Figure 32).
Colicins (MW > 10 kDa) are made mainly by Enterobacteriaceae (mainly E. coli) [44][32]. They can form pores in the cell wall or degrade bacteria nucleic acid structures (RNAses, DNAses, or tRNAses) [32][19]. Colicins can be grouped into four subclasses: colicins forming channels in the cytoplasmic membrane, colicins degrading DNA, colicins targeting rRNA or tRNA, and colicins inhibiting murein and lipopolysaccharide biosynthesis [45,46][33][34] (Figure 32).
Colicin-like bacteriocins are made by the Klebsiella genus (klebicins) and P. aeruginosa (S-type pyocins) [46][34]. They are similar in size, structure, and function to colicins.
Phage-tail-like bacteriocins have structures similar to phage tails. They are cylindrical peptides with high molecular weights [23][10]. They are grouped into the R-type and F-type subclasses [23][10] (Figure 32).
R-type phage-tail-like bacteriocins bind to cell surface receptors, force the internal core into the microbial cell envelope, and determine rapid cell death [23][10]. They also affect macromolecule synthesis and oxygen uptake [9][28]. F-type phage-tail-like bacteriocins have a mechanism of action similar to R-type, but do not have contractile movement [27][14].
3. Fungal AMPs
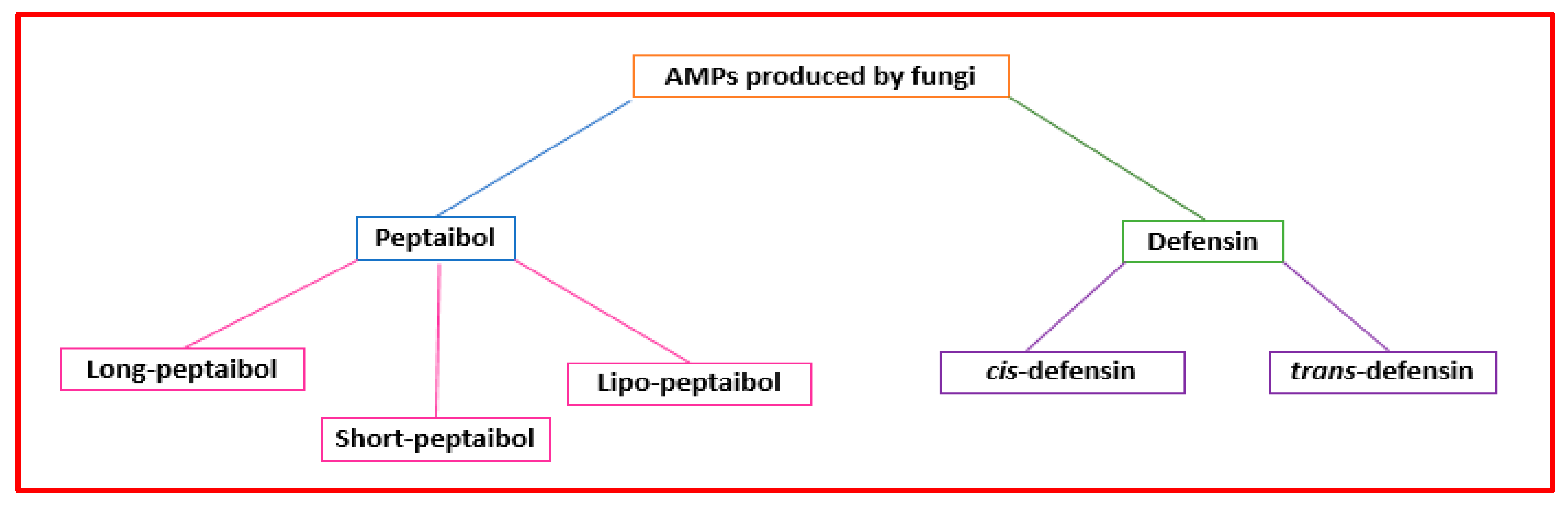
Figure 43.
Classification of the AMPs produced by fungi.
The term “peptaibol” is linked to structural characteristics. It is a combination of the words “peptide,” “α-aminoisobutyrate,” and “amino alcohol” [49][37]. Peptaibols are mainly made by Trichoderma fungi [50][38]. They are short peptides (containing 5–21 amino acids) with a high proportion of non-proteinogenic amino acids (i.e., α-aminoisobutyric acid), acylated N-terminal residue, and amino alcohol (i.e., leucenol or phenylalaninol) linked to the C-terminal [51][39]. Their three-dimensional structures consist of α-helix and β-bend patterns [52][40]. They are classified based on sequence length as “long” (18−20 residues), “short” (11−16 amino acids) (Figure 43), and founded on modification types on the terminal groups, “lipo” peptaibols (i.e., N-terminal acylated by decanoic) [53][41]. Different mechanisms have been proposed to describe their action. Concerning large peptaibols, it was hypothesized that their helical structures oligomerize and can form ion channels in the membrane. Instead, short peptaibols can form a pore via helical bundles (within the bilayer or by a barrel-stave mechanism) and interact with diverse molecular targets [9][28]. Peptaibols’ modes of action that do not involve interaction with the bacterial membrane include the inhibition of cell wall synthesis, DNA, protein synthesis, and that of relevant enzymes [10][42].
Eukaryotes and bacteria can produce defensins. Defensins are a class of cysteine-rich AMPs with short, cationic disulfide bridges [54][43]. They can be grouped into two superfamilies (cis and trans) (Figure 43). Fungi can produce cis-defensins with α-helical (cysteine-stabilized) or β-sheet folds. Defensins can disrupt the microbial cytoplasmic membrane, bind the bacterial precursor lipid II of the cell wall, or prevent cell wall biosynthesis [55][44].
4. Plant AMPs
Plant AMPs are the first line of defense against infections produced by pathogenic microorganisms. They can have diverse structures and action mechanisms. Their classification is based on their tridimensional structures and amino acid sequence similarity, including thionins, hevein-like peptides, defensins, knottins, stable-like peptides, snakins, lipid transfer proteins, and cyclotides [56][45] (Figure 54).
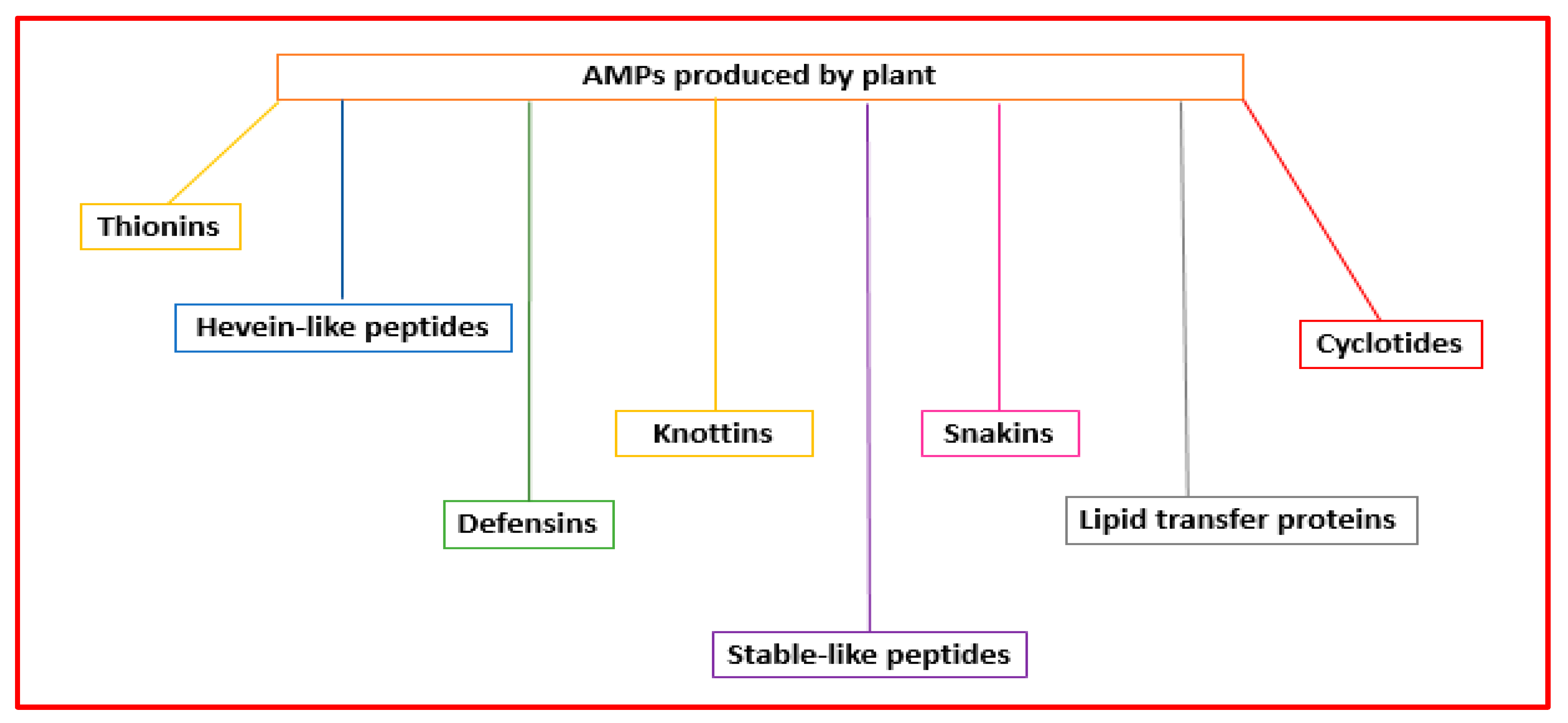
Figure 54.
Classification of the AMPs produced by plants.
4.1. Thionins
Thionins are classified into five types indicated by Roman numerals, have sizes ranging from 45 to 48, and are found in monocots and dicots. They include two distinct superfamilies: α/β-thionins and γ-thionins [57][46]. α/β thionins have similar structures (homologous amino acid sequences) [58][47] and are rich in arginine, cysteine, and lysine. γ-thionins are similar to defensins, so some authors classify them in this group [59][48]. Thionins have a broad spectrum of activities. They act against Gram-positive and Gram-negative bacteria, yeast, fungi, insect larvae, and nematodes [60,61,62][49][50][51] and present cytotoxic effects against mammal cells in vitro [63][52].
4.2. Hevein-like peptides
Hevein-like peptides can contain 29–45 amino acids with glycine (6), cysteine (8–10), and aromatic residues. They have a chitin-binding domain responsible for their antifungal activity [64][53] and 3–5 disulfide bonds that stabilize the antiparallel β-sheets and short α-helix [65][54]. The factors that favor chitin-binding are the three aromatic amino acids that give stability to the hydrophobic C-H group, the π electron system that determines van der Waals forces, and the hydrogen bonds between serine and N-acetylglucosamine [64][53]. Hevein-like peptides damage the fungal cell wall by interacting with hydrophobic residues and chitin present in the fungal cell [5][55]. They can constrain some enzymes’ activities by linking them with disulfide bonds [66][56].
4.3. Defensins
Defensins can comprise 45–54 amino acids and four disulfide bridges. They have an antiparallel β sheet, are enclosed by an α-helix, and are limited by intramolecular disulfide bonds [67][57] called cysteine-stabilized αβ (CSαβ) motifs [68][58]. Defensins are resistant to proteolysis and are stable to variations in temperature and pH. They prevent microbial growth, trypsin, and α-amylase activities, decrease abiotic stress, and change the redox state of ascorbic acid [56][45].
4.4. Knottins
Knottins, also called “cysteine-knot peptides”, are formed by 39 amino acids (of which six are cysteine residues), have three disulfide bonds (cysteine-knot motifs), and can be found in two conformations (cyclic and linear) [5,69][55][59]. They have high thermal stability and resistance to proteolytic action and can inhibit α-amylase, trypsin, carboxypeptidase, and cysteine protease [70,71][60][61]. They differ from protease inhibitors and defensins regarding cysteine space [5][55]. They are amphipathic peptides whose cationic portions can bind cell membranes, acid-sensing channels, and K+ and Na+ channels in membranes. Once they enter a cell, they attack intracellular targets (i.e., carboxypeptidases) and promote resistance [61][50]. Unfortunately, knottins are highly cytotoxic to human cells since their contact with membranes is not selective.
4.5. Stable-like Peptides
Stable-like peptides are a class of small peptides that form a helix-loop-helix structure with a typical Cys motif of XnC1X3C2XnC3X3C4Xn (-X is an amino acid residue different from cysteine). Although their amino acid sequence is highly variable, the three-dimensional structure of stable-like peptides is conserved. They can have antifungal, antibacterial, ribosome-inactivating, and trypsin inhibiting activities [72][62]. Their bacteriostatic effect is due to binding with DNA, which decreases RNA and protein synthesis [73][63]. Their activity relates to the loop region that connects the two α-helices [74][64].
4.6. Snakins
Snakins are generally small (~7 kDa), cysteine-rich, and positively charged proteins with antimicrobial, antinematode, and antifungal properties [75][65]. The mechanism of action is not precise. More than one hypothesis has been developed to explain it. Some authors believe they can promote immune responses by destabilizing the site of action through interaction with the negatively charged component [76,77][66][67]. Other authors hypothesized that they can act on phytohormone biosynthesis and transduction processes [78][68].
4.7. Lipid Transfer Proteins
Lipid transfer proteins (LTPs) are small, cysteine-rich proteins (containing 100 aa) having 4 to 5 helices in their structure that are stabilized by hydrogen bonds. They can transfer lipids (i.e., fatty acids, phospholipids, acyl CoA fatty acids, and sterols) between membranes. In this way, they form pores and determine cell death. They can be classified into two subfamilies, LTP1 (relative molecular weight of 9 kDa) and LTP2 (relative molecular weight of 7 kDa), or into five types (LTP1, LTP2, LTPc, LTPd, and LTPg) based on the position of the conserved intron, the space between the cysteine residues, and the identity of the amino acid sequence [69][59].
4.8. Cyclotides
Cyclotides are macrocyclic with cyclic cystine knot (CCK) structural motif peptides [79][69]. Disulfide bridges stabilize the head-to-tail cyclo. They can be classified into two subfamilies: Möbius and bracelets [80][70]. Their action depends on the cystine knot structural motif that promotes hydrophobic residue surface contact, some of which form a hydrophobic patch [81][71]. Cyclotides can act against bacteria, helminths, insects, and mollusks and have ecbolic anti-HIV and anticancer properties [81][71].
5. Animal AMPs
Vertebrate defensins are synthesized as “prepropeptides” and classified into α, β, and θ defensins [82][72]. They have short polypeptide sequences (18–45 amino acids), cationic net charges (+1 to +11), and three intramolecular disulfide bonds. In human α-defensins, the characteristic connections of disulfide bridges are Cys1–Cys6, Cys2–Cys4 and Cys3–Cys5 [83][73]. They are synthesized by promyelocytes and intestinal Paneth cells [84][74]. β-defensins differ from α-defensins in disulfide bond distributions and cysteine residues. The disulfide bridges in human β-defensins are Cys1–Cys5, Cys2–Cys4 and Cys3–Cys6 [83][73]. θ-defensins are cyclic octadecapeptides not expressed in humans and are active against B. anthrax, S. aureus, and C. albicans [85,86,87][75][76][77]. They contain a macrocyclic backbone and are structurally dissimilar to α- and β-defensins [88][78].
Invertebrates synthesize AMPs as components of humoral defense [89][79]. They are cationic peptides that can contain six or eight cysteine residues and show a cysteine-stabilized α/β motif [90][80]. The defensins produced by insects, arthropods, and mollusks contain six cysteines.
Invertebrate defensins are phylogenetically and structurally associated with vertebrate β-defensins. They have a hydrophobic domain (N-terminal) that can act against Gram-positive bacteria and a cationic domain (C-terminal containing six cysteines) that can act against Gram-negative bacteria [92][82].
Crustins (cationic cysteine-rich peptides that form a tightly packed structure) are found in crustaceans [93][83]. They have an N-terminal multidomain (rich in glycine, cysteine, and proline) and a C-terminal (with four C-terminal disulfide bridges) (Table 1) [94][84].
Table 1.
Animal AMPs.
| Animals | AMPs |
|---|---|
| Mammalians | cathelicidins defensins (α-, β-, and θ-defensins; θ-defensins are not expressed in adult humans) platelet antimicrobial proteins dermicidins hepcidins |
| Reptiles | defensins (α, β-, and θ-defensins) cathelicidins |
| Fish | β-defensins cathelicidins hepicidins (HAMP1 and HAMP2) histone-derived peptides piscidins (piscidins 1–7) |
| Amphibians | magainins cancrins |
| Crustaceans | crustins |
In fish, reptiles, amphibians, birds, and mammalians, AMPs ( size range of 15–200 residues) play an essential role in the immediate response to microorganisms [9][28]. Fish produce β-defensins, cathelicidins, hepicidins, histone-derived peptides, and piscidins [95][85] (Table 1).
Fish defensins are β-defensin-like proteins containing six cysteine motifs [96][86]. Cathelicidins are cationic proteins activated by elastase and other proteases discovered in the secretory granules of immune cells [97][87]. They act against Gram-positive and Gram-negative bacteria, parasites, fungi, and enveloped viruses [98,99,100,101,102][88][89][90][91][92]. Cathelicidins can bind and disrupt negatively charged membranes, alter RNA and DNA synthesis, damage the functions of enzymes and chaperones, and promote protein degradation [103][93].
Fish hepcidins are cysteine-rich peptides similar to human hepcidin with a hairpin structure linked via four disulfide bonds. They are iron-regulating antimicrobial hormones [104,105][94][95].
Hepcidins are grouped into HAMP1 and HAMP2 [95][85]. They act against bacteria (Gram-positive and Gram-negative) and fish pathogens and induce the internalization and degradation of ferroportin [106][96].
Piscidins are linear amphipathic AMPs. They have histidine residue and an α-helix that can interact with lipid bilayers [107][97]. They are classified into piscidins 1–7 based on their biological activity, amino acid sequence, and length [107][97].
References
- Danis-Wlodarczyk, K.M.; Wozniak, D.J.; Abedon, S.T. Treating Bacterial Infections with Bacteriophage-Based Enzybiotics: In Vitro, In Vivo and Clinical Application. Antibiotics 2021, 10, 1497.
- Nelson, D.; Loomis, L.; Fischetti, V.A. Prevention and elimination of upper respiratory colonization of mice by group A streptococci by using a bacteriophage lytic enzyme. Proc. Natl. Acad. Sci. USA 2001, 98, 4107–4112.
- Abdelrahman, F.; Easwaran, M.; Daramola, O.I.; Ragab, S.; Lynch, S.; Oduselu, T.J.; Khan, F.M.; Ayobami, A.; Adnan, F.; Torrents, E.; et al. Phage-Encoded Endolysins. Antibiotics 2021, 10, 124.
- Yan, J.; Mao, J.; Xie, J. Bacteriophage polysaccharide depolymerases and biomedical applications. BioDrugs 2014, 28, 265–274.
- Ha, E.; Son, B.; Ryu, S. Clostridium perfringens virulent bacteriophage CPS2 and its thermostable endolysin lysCPS2. Viruses 2018, 10, 251.
- Plotka, M.; Kapusta, M.; Dorawa, S.; Kaczorowska, A.K.; Kaczorowski, T. Ts2631 endolysin from the extremophilic thermus scotoductus bacteriophage vB_Tsc2631 as an antimicrobial agent against gram-negative multidrug-resistant bacteria. Viruses 2019, 11, 657.
- Pastagia, M.; Schuch, R.; Fischetti, V.A.; Huang, D.B. Lysins: The arrival of pathogen-directed anti-infectives. J. Med. Microbiol. 2013, 62, 1506–1516.
- Abril, A.G.; Carrera, M.; Notario, V.; Sánchez-Pérez, Á.; Villa, T.G. The Use of Bacteriophages in Biotechnology and Recent Insights into Proteomics. Antibiotics 2022, 11, 653.
- Latka, A.; Maciejewska, B.; Majkowska-Skrobek, G.; Briers, Y.; Drulis-Kawa, Z. Bacteriophage-encoded virion-associated enzymes to overcome the carbohydrate barriers during the infection process. Appl. Microbiol. Biotechnol. 2017, 101, 3103–3119.
- Scholl, D. Phage Tail-Like Bacteriocins. Annu. Rev. Virol. 2017, 4, 453–467.
- Daw, M.A.; Falkiner, F.R. Bacteriocins: Nature, function and Structure. Micron 1996, 27, 467–479.
- Chen, J.; Zhu, Y.; Yin, M.; Xu, Y.; Liang, X.; Huang, Y.P. Characterization of maltocin S16, a phage tail-like bacteriocin with antibacterial activity against Stenotrophomonas maltophilia and Escherichia coli. J. Appl. Microbiol. 2019, 127, 78–87.
- Morse, S.A.; Jones, B.V.; Lysko, P.G. Pyocin inhibition of Neisseria gonorrhoea: Mechanism of action. Antimicrob. Agents Chemother. 1980, 18, 416–423.
- Lee, G.; Chakraborty, U.; Gebhart, D.; Govoni, G.R.; Zhou, Z.H.; Scholl, D. F-type bacteriocins of Listeria monocytogenes: A new class of phage tail-like structures reveals broad parallel coevolution between tailed bacteriophages and high-molecular-weight bacteriocins. J. Bacteriol. 2016, 198, 2784–2793.
- Tajbakhsh, M.; Karimi, A.; Fallah, F.; Akhavan, M.M. Overview of ribosomal and non-ribosomal antimicrobial peptides produced by Gram positive bacteria. Cell. Mol. Biol. 2017, 6, 20.
- Diep, D.; Nes, I. Ribosomally Synthesized Antibacterial Peptides in Gram Positive Bacteria. Curr. Drug Targets 2005, 3, 107–122.
- Yeaman, M.R.; Yount, N.Y. Mechanisms of antimicrobial peptide action and resistance. Pharmacol. Rev. 2003, 55, 27–55.
- Kumariya, R.; Kumari, G.; Raiput, Y.S.; Akhtar, N.; Patel, S. Bacteriocins: Classification, synthesis, mechanism of action and resistance development in food spoilage causing bacteria. Microb. Pathog. 2019, 128, 171–177.
- Meade, E.; Slattery, M.A.; Garvey, M. Bacteriocins, Potent Antimicrobial Peptides and the Fight against Multi Drug Resistant Species: Resistance Is Futile? Antibiotics 2020, 9, 32.
- Gradisteanu Pircalabioru, G.; Popa, L.I.; Marutescu, L.; Gheorghe, I.; Popa, M.; Czobor Barbu, I.; Cristescu, R.; Chifiriuc, M.-C. Bacteriocins in the Era of Antibiotic Resistance: Rising to the Challenge. Pharmaceutics 2021, 13, 196.
- Bierbaum, G.; Sahl, H.-G. Lantibiotics: Mode of Action, Biosynthesis and Bioengineering. Curr. Pharm. Biotechnol. 2009, 10, 2–18.
- Cotter, P.D.; Hill, C.; Ross, R.P. Food microbiology: Bacteriocins: Developing innate immunity for food. Nat. Rev. Microbiol. 2005, 3, 777–788.
- Cintas, L.M.; Casaus, P.; Håvarstein, L.S.; Hernández, P.E.; Nes, I.F. Biochemical and genetic characterization of enterocin P, a novel sec-dependent bacteriocin from Enterococcus faecium P13 with a broad antimicrobial spectrum. Appl. Environ. Microbiol. 1997, 63, 4321–4330.
- Nissen-Meyer, J.; Oppegård, C.; Rogne, P.; Haugen, H.S.; Kristiansen, P.E. Structure and mode-of-action of the two-peptide (class-IIb) bacteriocins. Probiotics Antimicrob. Proteins 2010, 2, 52–60.
- Van Belkum, M.J.; Martin-Visscher, L.A.; Vederas, J.C. Structure and genetics of circular bacteriocins. Trends Microbiol. 2011, 19, 411–418.
- Nissen-Meyer, J.; Rogne, P.; Oppegard, C.; Haugen, H.; Kristiansen, P. Structure-Function Relationships of the Non-Lanthionine-Containing Peptide (class II) Bacteriocins Produced by Gram-Positive Bacteria. Curr. Pharm. Biotechnol. 2009, 10, 19–37.
- Heng, N.C.K.; Wescombe, P.A.; Burton, J.P.; Jack, R.W.; Tagg, J.R. The diversity of bacteriocins in Gram-positive bacteria. In Bacteriocins: Ecology and Evolution; Riley, M.A., Chavan, M.A., Eds.; Springer: New York, NY, USA, 2007; pp. 45–92.
- Bin Hafeez, A.; Jiang, X.; Bergen, P.J.; Zhu, Y. Antimicrobial Peptides: An Update on Classifications and Databases. Int. J. Mol. Sci. 2021, 22, 11691.
- Simons, A.; Alhanout, K.; Duval, R.E. Bacteriocins, Antimicrobial Peptides from Bacterial Origin: Overview of Their Biology and Their Impact against Multidrug-Resistant Bacteria. Microorganisms 2020, 8, 639.
- Duquesne, S.; Destoumieux-Garzón, D.; Peduzzi, J.; Rebuffat, S. Microcins, gene-encoded antibacterial peptides from enterobacteria. Nat. Prod. Rep. 2007, 24, 708–734.
- Pons, A.M.; Lanneluc, I.; Cottenceau, G.; Sable, S. New developments in non-post translationally modified microcins. Biochimie 2002, 84, 531–537.
- Cascales, E.; Buchanan, S.K.; Duche, D.; Kleanthous, C.; Lloubes, R.; Postle, K.; Riley, M.; Slatin, S.; Cavard, D. Colicin biology. Microbiol. Mol. Biol. Rev. 2007, 71, 158–229.
- Gillor, O.; Kirkup, B.C.; Riley, M.A. Colicins and microcins: The next generation antimicrobials. Adv. Appl. Microbiol. 2004, 54, 129–146.
- Papadakos, G.; Wojdyla, J.A.; Kleanthous, C. Nuclease colicins and their immunity proteins. Q. Rev. Biophys. 2012, 45, 57–103.
- Duclohier, H. Antimicrobial Peptides and Peptaibols, Substitutes for Conventional Antibiotics. Curr. Pharm. Des. 2010, 16, 3212–3223.
- Wu, J.; Gao, B.; Zhu, S. The fungal defensin family enlarged. Pharmaceuticals 2014, 7, 866–880.
- Evans, B.S.; Robinson, S.J.; Kelleher, N.L. Surveys of non-ribosomal peptide and polyketide assembly lines in fungi and prospects for their analysis in vitro and in vivo. Fungal Genet. Biol. 2011, 48, 49–61.
- Tyśkiewicz, R.; Nowak, A.; Ozimek, E.; Jaroszuk-Ściseł, J. Trichoderma: The Current Status of Its Application in Agriculture for the Biocontrol of Fungal Phytopathogens and Stimulation of Plant Growth. Int. J. Mol. Sci. 2022, 23, 2329.
- Leitgeb, B.; Szekeres, A.; Manczinger, L.; Vágvölgyi, C.; Kredics, L. The history of Alamethicin: A review of the most extensively studied peptaibol. Chem. Biodivers. 2007, 4, 1027–1051.
- Szekeres, A.; Leiteb, B.; Kredics, L.; Antal, Z.; Hatvani, L.; Manczinger, L.; Vágvölgyi, C. Peptaibols and related peptaibiotics of Trichoderma. Acta Microbiol. Immunol. Hung. 2005, 52, 137–168.
- Hou, X.; Sun, R.; Feng, Y.; Zhang, R.; Zhu, T.; Che, Q.; Zhang, G.; Li, D. Peptaibols: Diversity, bioactivity, and biosynthesis. Eng. Microbiol. 2022, 2, 100026.
- Wang, C.K.; Craik, D.J. Designing macrocyclic disulfide-rich peptides for biotechnological applications perspective. Nat. Chem. Biol. 2018, 14, 417–427.
- Qi, S.; Gao, B.; Zhu, S. A Fungal Defensin Inhibiting Bacterial Cell-Wall Biosynthesis with Non-Hemolysis and Serum Stability. J. Fungi 2022, 8, 174.
- Schneider, T.; Kruse, T.; Wimmer, R.; Wiedemann, I.; Sass, V.; Pag, U.; Jansen, A.; Nielsen, A.K.; Mygind, P.H.; Raventós, D.S.; et al. Plectasin, a fungal defensin, targets the bacterial cell wall precursor lipid II. Science 2010, 328, 1168–1172.
- Lima, A.M.; Azevedo, M.I.; Sousa, L.M.; Oliveira, N.S.; Andrade, C.R.; Freitas, C.D.; Souza, P.F. Plant antimicrobial peptides: An overview about classification, toxicity and clinical applications. Int. J. Biol. Macromol. 2022, 214, 10–21.
- Goyal, R.K.; Mattoo, A.K. Plant antimicrobial peptides. In Host Defense Peptides and Their Potential as Therapeutic Agents; Epand, R.M., Ed.; Springer: Cham, Switzerland, 2016; pp. 111–136.
- Thevissen, K.; Ferket, K.K.A.; François, I.E.J.A.; Cammue, B.P.A. Interactions of antifungal plant defensins with fungal membrane components. Peptides 2003, 24, 1705–1712.
- Paul, M.; Chowdhury, T.; Saha, S. Antimicrobial peptide: A competent tool for plant disease control in mulberry—A review. Vegetos 2022, 1–10.
- Vasilchenko, A.S.; Smirnov, A.N.; Zavriev, S.K.; Grishin, E.V.; Vasilchenko, A.V.; Rogozhin, E.A. Novel thionins from black seed (Nigella sativa L.) demonstrate antimicrobial activity. Int. J. Pept. Res. Ther. 2017, 23, 171–180.
- Moore, S.J.; Leung, C.L.; Cochran, J.R. Knottins: Disulfide-bonded therapeutic and diagnostic peptides. Drug Discov. Today Technol. 2012, 9, e3–e11.
- Taveira, G.B.; Carvalho, A.O.; Rodrigues, R.; Trindade, F.G.; Da Cunha, M.; Gomes, V.M. Thionin-like peptide from Capsicum annuum fruits: Mechanism of action and synergism with fluconazole against Candida species. BMC Microbiol. 2016, 16, 12.
- Taveira, G.B.; Mello, É.O.; Carvalho, A.O.; Regente, M.; Pinedo, M.; de La Canal, L.; Rodrigues, R.; Gomes, V.M. Antimicrobial activity and mechanism of action of a thionin-like peptide from Capsicum annuum fruits and combinatorial treatment with fluconazole against Fusarium solani. Biopolymers 2017, 108, e23008.
- Slavokhotova, A.A.; Shelenkov, A.A.; Andreev, Y.A.; Odintsova, T.I. Hevein-Like Antimicrobial Peptides of Plants. Biochemistry 2017, 82, 1659–1674.
- Odintsova, T.; Shcherbakova, L.; Slezina, M.; Pasechnik, T.; Kartabaeva, B.; Istomina, E.; Dzhavakhiya, V. Hevein-like antimicrobial peptides WAMPs: Structure-function relationship in antifungal activity and sensitization of plant pathogenic fungi to tebuconazole by WAMP-2-derived peptides. Int. J. Mol. Sci. 2020, 21, 7912.
- Tam, J.P.; Wang, S.; Wong, K.H.; Tan, W.L. Antimicrobial Peptides from Plants. Pharmaceuticals 2015, 8, 711–757.
- Kramer, K.J.; Klassen, L.W.; Jones, B.L.; Speirs, R.D.; Kammer, A.E. Toxicity of purothionin and its homologues to the tobacco hornworm, Manduca sexta (L.) (lepidoptera:Sphingidae). Toxicol. Appl. Pharmacol. 1979, 48, 179–183.
- Azmi, S.; Hussain, M.K. Analysis of structures, functions, and transgenicity of phytopeptides defensin and thionin: A review. Beni Suef. Univ. J. Basic Appl. Sci. 2021, 10, 5.
- Gao, B.; Zhu, S. A Fungal Defensin Targets the SARS−CoV−2 Spike Receptor−Binding Domain. J. Fungi 2021, 7, 553.
- dos Santos-Silva, C.A.; Zupin, L.; Oliveira-Lima, M.; Vilela, L.M.B.; Bezerra-Neto, J.P.; Ferreira-Neto, J.R.; Ferreira, J.D.C.; de Oliveira-Silva, R.L.; de Pires, C.J.; Aburjaile, F.F.; et al. Plant Antimicrobial Peptides: State of the Art, In Silico Prediction and Perspectives in the Omics Era. Bioinform. Biol. Insights 2020, 14, 117793222095273.
- Hellinger, R.; Gruber, C.W. Peptide-based protease inhibitors from plants. Drug Discov. Today 2019, 24, 1877–1889.
- Molesini, B.; Treggiari, D.; Dalbeni, A.; Minuz, P.; Pandolfini, T. Plant cystine-knot peptides: Pharmacological perspectives. Br. J. Clin. Pharmacol. 2017, 83, 63–70.
- Postic, G.; Gracy, J.; Périn, C.; Chiche, L.; Gelly, J.-C. KNOTTIN: The database of inhibitor cystine knot scaffold after 10 years, toward a systematic structure modeling. Nucleic Acids Res. 2017, 46, D454–D458.
- Slavokhotova, A.A.; Rogozhin, E.A. Defense Peptides From the α-Hairpinin Family Are Components of Plant Innate Immunity. Front. Plant Sci. 2020, 11, 465.
- Haney, E.F.; Petersen, A.P.; Lau, C.K.; Jing, W.; Storey, D.G.; Vogel, H.J. Mechanism of action of puroindoline derived tryptophan-rich antimicrobial peptides. Biochim. Biophys. Acta (BBA) Biomembr. 2013, 1828, 1802–1813.
- Rogozhin, E.; Ryazantsev, D.; Smirnov, A.; Zavriev, S. Primary Structure Analysis of Antifungal Peptides from Cultivated and Wild Cereals. Plants 2018, 7, 74.
- Su, T.; Han, M.; Cao, D.; Xu, M. Molecular and Biological Properties of Snakins: The Foremost Cysteine-Rich Plant Host Defense Peptides. J. Fungi 2020, 6, 220.
- Yeung, H.; Squire, C.J.; Yosaatmadja, Y.; Panjikar, S.; López, G.; Molina, A.; Baker, E.N.; Harris, P.W.; Brimble, M.A. Protein Structures Very Important Paper Radiation Damage and Racemic Protein Crystallography Reveal the Unique Structure of the GASA/Snakin Protein Superfamily. Angew. Chem. 2016, 128, 8062–8065.
- Rodríguez, S.; Mariana, D.; Vega, B.; Dans, P.D.; Pandolfi, V.; Benko-Iseppon, A.M.; Cecchetto, G. Antimicrobial and structural insights of a new snakin-like peptide isolated from Peltophorum dubium (Fabaceae). Amino Acids 2018, 50, 1245–1259.
- Zhang, S.; Wang, X. One new kind of phytohormonal signaling integrator: Up-and-coming GASA family genes. Plant Signal. Behav. 2017, 12, e1226453.
- de Veer, S.J.; Kan, M.-W.; Craik, D.J. Cyclotides: From Structure to Function. Chem. Rev. 2019, 119, 12375–12421.
- Carla Barbosa da Silva Lima, S.; Maria Benko-Iseppon, A.; Pacifico Bezerra Neto, J.; Lindinalva Barbosa Amorim, L.; Ribamar Costa Ferreira Neto, J.; Crovella, S.; Pandolfi, V. Plants defense-related cyclic peptides: Diversity, structure and applications. Curr Protein Pept Sci. 2017, 18, 375–390.
- Huang, Y.-H.; Du, Q.; Craik, D.J. Cyclotides: Disulfide-rich peptide toxins in plants. Toxicon 2019, 172, 33–44.
- Selsted, M.E.; Ouellette, A.J. Mammalian defensins in the antimicrobial immune response. Nat. Immunol. 2005, 6, 551–557.
- Niyonsaba, F.; Nagaoka, I.; Ogawa, H.; Okumura, K. Multifunctional antimicrobial proteins and peptides: Natural activators of immune systems. Curr. Pharm. Des. 2009, 15, 2393–2413.
- Ayabe, T.; Satchell, D.P.; Wilson, C.L.; Parks, W.C.; Selsted, M.E.; Ouellette, A.J. Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat. Immunol. 2000, 1, 113–118.
- Basso, V.; Garcia, A.; Tran, D.Q.; Schaal, J.B.; Tran, P.; Ngole, D.; Aqeel, Y.; Tongaonkar, P.; Ouellette, A.J.; Selsteda, M.E. Fungicidal Potency and Mechanisms of –Defensins against Multidrug-Resistant Candida Species. Antimicrob. Agents Chemother. 2018, 62, e00111-18.
- Wilmes, M.; Stockem, M.; Bierbaum, G.; Schlag, M.; Götz, F.; Tran, D.Q.; Schaal, J.B.; Ouellette, A.J.; Selsted, M.E.; Sahl, H.-G. Killing of Staphylococci by θ-Defensins Involves Membrane Impairment and Activation of Autolytic Enzymes. Antibiotics 2014, 3, 617–631.
- Welkos, S.; Cote, C.K.; Hahn, U.; Shastak, O.; Jedermann, J.; Bozue, J.; Jung, G.; Ruchala, P.; Pratikhya, P.; Tang, T.; et al. Humanized theta-defensins (retrocyclins) enhance macrophage performance and protect mice from experimental anthrax infections. Antimicrob. Agents Chemother. 2011, 55, 4238–4250.
- Hazlett, L.; Wu, M. Defensins in innate immunity. Cell Tissue Res. 2011, 343, 175–188.
- Rowley, A.F.; Powell, A. Invertebrate immune systems specific, quasi-specific, or nonspecific? J. Immunol. 2007, 179, 7209–7214.
- Froy, O. Convergent evolution of invertebrate defensins and nematode antibacterial factors. Trends Microbiol. 2005, 13, 314–319.
- Tassanakajon, A.; Somboonwiwat, K.; Amparyup, P. Sequence diversity and evolution of antimicrobial peptides in invertebrates. Dev. Comp. Immunol. 2015, 48, 324–341.
- Saito, T.; Kawabata, S.I.; Shigenaga, T.; Takayenoki, Y.; Cho, J.; Nakajima, H.; Hirata, M.; Iwanaga, S. A novel big defensin identified in horseshoe crab hemocytes: Isolation, amino acid sequence, and antibacterial activity. J. Biochem. 1995, 117, 1131–1137.
- Ranganathan, S.; Simpson, K.J.; Shaw, D.C.; Nicholas, K.R. The whey acidic protein family: A new signature motif and three-dimensional structure by comparative modeling. J. Mol. Graph. Model. 1999, 17, 106–113.
- Smith, V.J. Phylogeny of whey acidic protein (WAP) four-disulfide core proteins and their role in lower vertebrates and invertebrates. Biochem. Soc. Trans. 2011, 39, 1403–1408.
- Masso-Silva, J.A.; Diamond, G. Antimicrobial peptides from fish. Pharmaceuticals 2014, 7, 265–310.
- Nam, B.H.; Moon, J.Y.; Kim, Y.O.; Kong, H.J.; Kim, W.J.; Lee, S.J.; Kim, K.K. Multiple β-defensin isoforms identified in early developmental stages of the teleost Paralichthys olivaceus. Fish Shellfish Immunol. 2010, 28, 267–274.
- Van Harten, R.M.; Van Woudenbergh, E.; Van Dijk, A.; Haagsman, H.P. Cathelicidins: Immunomodulatory Antimicrobials. Vaccines 2018, 6, 63.
- Goitsuka, R.; Chen, C.-L.H.; Benyon, L.; Asano, Y.; Kitamura, D.; Cooper, M.D. Chicken cathelicidin-b1, an antimicrobial guardian at the mucosal m cell gateway. Proc. Natl. Acad. Sci. USA 2007, 104, 15063–15068.
- Tossi, A.; Scocchi, M.; Zanetti, M.; Storici, P.; Gennaro, R. Pmap-37, a novel antibacterial peptide from pig myeloid cells. Cdna cloning, chemical synthesis and activity. Eur. J. Biochem. 1995, 228, 941–946.
- Veldhuizen, E.J.A.; Scheenstra, M.R.; Tjeerdsma-van Bokhoven, J.L.M.; Coorens, M.; Schneider, V.A.F.; Bikker, F.J.; van Dijk, A.; Haagsman, H.P. Antimicrobial and immunomodulatory activity of pmap-23 derived peptides. Protein Pept. Lett. 2017, 24, 609–616.
- Wessely-Szponder, J.; Majer-Dziedzic, B.; Smolira, A. Analysis of antimicrobial peptides from porcine neutrophils. J. Microbiol. Methods 2010, 83, 8–12.
- Xiao, Y.; Cai, Y.; Bommineni, Y.R.; Fernando, S.C.; Prakash, O.; Gilliland, S.E.; Zhang, G. Identification and functional characterization of three chicken cathelicidins with potent antimicrobial activity. J. Biol. Chem. 2006, 281, 2858–2867.
- Liu, Z.-M.; Chen, J.; Lv, Y.-P.; Hu, Z.-H.; Dai, Q.-M.; Fan, X.-L. Molecular characterization of a hepcidin homologue in starry flounder (Platichthys stellatus) and its synergistic interaction with antibiotics. Fish Shellfish Immunol. 2018, 83, 45–51.
- Huang, P.H.; Chen, J.Y.; Kuo, C.M. Three different hepcidins from tilapia, Oreochromis mossambicus: Analysis of their expressions and biological functions. Mol. Immunol. 2007, 44, 1922–1934.
- Hunter, H.N.; Bruce Fulton, D.; Ganz, T.; Vogel, H.J. The solution structure of human hepcidin, a peptide hormone with antimicrobial activity that is involved in iron uptake and hereditary hemochromatosis. J. Biol. Chem. 2002, 277, 37597–37603.
- Chaturvedi, P.; Bhat, R.A.H.; Pande, A. Antimicrobial Peptides of Fish: Innocuous Alternatives to Antibiotics. Rev. Aquac. 2020, 12, 85–106.
- Mihailescu, M.; Sorci, M.; Seckute, J.; Silin, V.I.; Hammer, J.; Perrin, P.S., Jr.; Hernandez, J.I.; Smajic, N.; Shrestha, A.; Bogardus, K.A.; et al. structure and function in antimicrobial piscidins: Histidine position, directionality of membrane insertion, and pH-dependent permeabilization. J. Am. Chem. Soc. 2019, 141, 9837–9853.
- Van Hoek, ML Antimicrobial Peptides in Reptiles. Pharmaceuticals 2014, 7, 723–753.
- Huan, Y.; Kong, Q.; Mou, H.; Yi, H. Antimicrobial Peptides: Classification, Design, Application and Research Progress in Multiple Fields. Front. Microbiol. 2020, 11, 582779.
- Cheng, Y.; Prickett, M.D.; Gutowska, W.; Kuo, R.; Belov, K.; Burt, D.W. Evolution of the avian β-defensin and cathelicidin genes. BMC Evol. Biol. 2015, 15, 188.
More
