Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Qingzhong Wang and Version 2 by Beatrix Zheng.
Viral DNA and RNA polymerases are two kinds of very important enzymes that synthesize the genetic materials of the virus itself, and they have become extremely favorable targets for the development of antiviral drugs because of their relatively conserved characteristics. There are many similarities in the structure and function of different viral polymerases, so inhibitors designed for a certain viral polymerase have acted as effective universal inhibitors on other types of viruses.
- viral DNA and RNA polymerase
- antiviral drugs
- inhibitors
1. Viral DNA/RNA Synthases Inhibitors
After the virus enters the host cell, the host cell is used to synthesize the virus’ own nucleic acid and protein [1][47]. Most of the antiviral drugs that are clinically used act on the viral replication stage [2][48].
1.1. Influenza Virus RNA Polymerase Inhibitor
The replication for most viral RdRp is processed within the host cells. Influenza virus RdRp consists of three subunits encoded by the virus: PB1, PB2, and PA [3][49]. The naked viral genomic RNA must be combined with nucleic proteins to form a complex that serves as a template to initiate viral genome replication and transcription by RdRp. Studies have shown that the three subunits of influenza virus RdRp can enter the nucleus individually [4][5][6][50,51,52]. First, RdRp is transported by the nuclear localization signal (NLS) of the nucleoprotein (NP) into the host’s nucleus for assembly [7][53]. After assembly, the replication process of the influenza virus’ genetic material begins. Since the influenza virus cannot produce 5’-cap primers on its own, its PB2 subunit captures the 5’-cap structure of host cell RNA through a “cap capture” mechanism [8][54]. After replication is completed, the product is exported through a separate channel for viral mRNA synthesis [9][55]. Different types of inhibitors targeting the PB1, PB2, and PA subunits are discussed in detail below [10][11][56,57].
1.1.1. PA Inhibitors
At present, the PA inhibitor baloxavir marboxil, jointly developed by Shionogi and Roche, is used for the treatment of influenza A and B in individuals over the age of 12 [12][58]. Since there is no similar mechanism and corresponding protease in host cells, baloxavir acts as a novel CAP-dependent nucleic acid endonuclease inhibitor, and can selectively block the transcription process of the influenza virus without affecting host cells [13][14][59,60]. The results of clinical trials show that baloxavir can significantly improve the time of symptom relief and that this drug is well-tolerated [15][61].
1.1.2. PB1 Inhibitors
Among the PB1 inhibitors, ribavirin and favipiravir have entered the clinical research stage due to their good antiviral activity [16][62]. Although both compounds contain bases in their structures, their antiviral mechanisms are not the same [17][63]. Ribavirin is a broad-spectrum antiviral drug widely accepted as a competitive inhibitor of host monophosphate dehydrogenase (IMPDH) [18][64]. IMPDH can catalyze the conversion of guanosine monophosphate (GMP) to guanine triphosphate (GTP), and inhibition of this enzyme will reduce the content of GTP in cells, resulting in an imbalance in nucleotide concentration, thereby inhibiting viral protein synthesis and exerting an antiviral effect [19][65]. Favipiravir belongs to a class of purine analogs that need to be rapidly converted into triphosphate form in vivo, and it exerts an antiviral effect by simulating competitive inhibition of RNA polymerase activity by GTP [20][21][66,67]. At the same time, it can also be incorporated into viral genes and exert antiviral effects by inducing lethal mutations [22][68]. In addition to being anti-influenza virus, favipiravir is effective against a variety of RNA viruses such as Lassa fever virus, Rift Valley fever virus, Hantavirus, Flaviviridae, West Nile virus, Zika virus, Chikungunya virus, Ebola virus, etc [20][66].
1.1.3. PB2 Inhibitors
Pimodivir, also known as VX-787, is a typical representative of the PB2 cap-binding domain inhibitors [23][69]. Based on phase III clinical study data, pimodivir did not show better efficacy than the existing standard compound, so Janssen decided to stop its clinical development as an anti-influenza A virus drug [24][70].
1.1.4. Protein–Protein Interaction Inhibitors
The three subunits of RdRp are non-covalently combined into a functional complex, so blocking the interaction between the subunits can effectively inhibit the activity of RdRp [25][71]. The polymerase inhibitors developed based on this mechanism are called protein–protein interaction inhibitors (PPI inhibitors) [26][72]. At present, the most commonly studied PPI inhibitors are PA-PB1 inhibitors. Massari et al. (2013) found that cycloheptathiophene-3-carboxamide compounds had weak PA-PB1 inhibitory activity but no antiviral activity through ELISA experiments, and further modified cycloheptathiophene-3-carboxamide compounds by considering their structure–activity relationship. They also synthesized 35 compounds, of which 1 and 2 had the strongest activity and whose structures are shown in Figure 1; their IC50 values were 32 µmol/L and 35 µmol/L, respectively, and they had no cytotoxicity at the concentration of 250 µmol/L. These two compounds became potential PPI inhibitors [27][73].
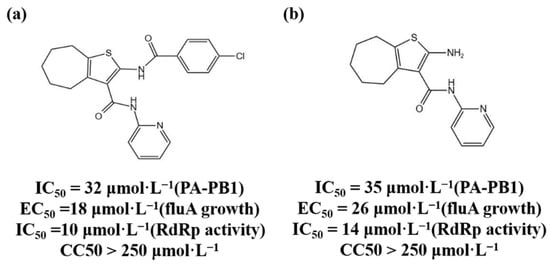
Figure 1.
(
a
,
b
) Chemical structures of Protein–protein interaction inhibitors and their antiviral activities.
1.2. HCV-related Inhibitors
The NS5B polymerase encoded by the non-structural gene NS5B is an enzyme of RdRp [28][74]. Its structure has the “right-hand” conformation of a typical RNA polymerase. The “palm subdomain“ is the central part of the polymerase and is the catalytic center of genome replication; the “fingers subdomain“ is responsible for capturing the nucleotide triphosphates required for replication; and the “thumb subdomain“ coordinates the initiation and elongation of RNA replication [29][75]. HCV replicates by using the viral genome’s single-stranded RNA as a template and joining the replicon created by the NS5B polymerase [30][76]. Because human cells lack similar enzymes, NS5B polymerase is a promising antiviral drug target. HCV NS5B polymerase inhibitors are divided into two categories: NIs and non-nucleoside inhibitors (NNIs) [31][77].
1.2.1. NIs
Sofosbuvir is a uridine analog prodrug, which can specifically inhibit HCV NS5B polymerase activity and is used for antiviral therapy in patients with chronic hepatitis C. The mechanism of action involves sofosbuvir being converted into the active uridine triphosphate under the action of intracellular phosphokinase, which competes with intracellular nucleotide phosphate as a substrate for NS5B polymerase and is incorporated into newly synthesized RNA chain, terminating the elongation of the HCV RNA chain, thereby inhibiting the replication of HCV [32][78]. Sofosbuvir has no inhibitory activity on human DNA and RNA polymerase, nor on mitochondrial RNA polymerase, so it has strong specificity [33][79]. Meanwhile, Sofosbuvir is a “pan-genotype” anti-HCV drug which not only inhibits HCV genotype 1 disease but is also effective against other genotypes of HCV infection.
1.2.2. NNIs
NNIs bind to the allosteric site of NS5B polymerase, resulting in a change in enzyme conformation, thereby inhibiting the activity of NS5B polymerase and exerting an antiviral effect [34][35][80,81]. Compared with NIs, NNIs have a lower genetic barrier and are prone to drug resistance and relapse after drug withdrawal, and cannot have antiviral effects on all genotypes [36][82]. Among them, dasabuvir sodium hydrate was approved by the European Medicines Agency (EMA) in January 2015 as an NNI-type drug.
1.2.3. NS5A Inhibitors
The HCV nonstructural protein NS5A is one of the components of the viral RNA replication complex [37][83]. It has not yet been found to have enzymatic activity, but it is essential for HCV RNA replication and is also related to the INF response [38][84]. NS5A inhibitors may exert anti-HCV effects by inhibiting the hyperphosphorylation of NS5A or altering the subcellular localization of NS5A [39][85]. Daclatasvir is a representative NS5A inhibitors which has a strong antiviral effect and is a pan-genotype drug [40][86].
1.3. HIV-1 Reverse Transcriptase Inhibitors
After retroviruses such as HIV-1 enter cells, they first synthesize RNA-DNA complex strands under the action of reverse transcriptase, and then the RNA strands are hydrolyzed to synthesize double-stranded DNA [41][87]. Under the action of the enzyme, this DNA is embedded into the genome of the host cell and replicated with the transcription and translation machinery of the host cell [42][88]. HIV-1 RT is the product of HIV-1 pol gene and consists of p51 and p66 subunits; the RT active center is located in the p66 subunit, which has RNA-dependent DNA polymerase activity [41][43][87,89]. Its DNA polymerase domain also has a “right-hand” conformation, similar to the RNA virus polymerase. It has four subdomains: the finger, thumb, palm, and linker domains. It is mainly responsible for the synthesis of double-stranded DNA using HIV single-stranded RNA as a template. The finger, thumb, and palm subdomains together constitute the active site, which occurs so that the enzyme is able to locate the template-primer accurately, and the palm domain contains the key site of DNA chain replication and elongation [44][90]. There are currently two types of reverse transcriptase inhibitors: NRTIs and NNRTIs [45][91].
1.3.1. NRTIs
NRTIs are analogs of deoxynucleotides, the DNA RT substrates for the synthesis of HIV-1. In vivo, NRTIs are converted into active nucleoside triphosphate derivatives, which compete with the natural deoxynucleoside triphosphate to bind HIV-1 RT to inhibit RT activity or terminate the RNA chain [46][47][92,93]. Currently approved NRTIs include zidovudine (AZT), didanosine (ddI), zalcitabine (ddC), stanvudine (d4T), lamivudine (3TC), abacavir (ABC), emtricitabine [(-)FTC], tenofovir (TFV), etc. [48][94].
1.3.2. NNRTIs
NNRTIs can directly combine with the p66 hydrophobic region of HIV-1 RT to change the conformation of the enzyme protein and inhibit the enzyme’s activity [41][49][87,95]. Unlike NRTIs, NNRTIs are not directly incorporated into DNA strands and do not require phosphorylation, and can function in quiescent and activated cells [50][96]. NNRTIs currently approved for clinical use include nevirapine (NVP), delavirdine (DLV), efavirenz (EFV), etravirine (TMC-125), and rilpivirine (RPV) [51][97].
1.3.3. HIV-1 Integrase Inhibitors
HIV-1 integrase catalyzes the integration of the HIV-1 virus reverse transcription product cDNA into the human host genome [52][98]. HIV-1 integrase is encoded by the 3’-end of the viral pol gene and contains 288 amino acid residues [53][99]. The integrase has 3’ cleavage endonuclease activity or strand transfer activity in vivo [54][100]. Inhibition of the HIV-1 integration process can be achieved by inhibiting the endonuclease activity or strand transfer activity of the integrase. There are currently four HIV-1 integrase inhibitors approved for clinical mediation, namely raltegravir, elvitegravir, dolutegravir, and bictegravir, and all four drugs act on the chain transfer process [55][101].
1.3.4. SARS-CoV-2 Enzyme Inhibitors
The structure of the polymerase complex of SARS-CoV-2 includes one NSP12, one NSP7, and two NSP8 subunits, which are required to complete the replication process of the coronavirus RNA [56][102]. The core catalytic site of the coronavirus polymerase is NSP12, and its RdRp domain is present in the standard “right-hand” conformation, including the three finger, thumb, and palm subdomains [57][103]. The RdRp associates with additional non-structural proteins to form a replication-transcription complex that carries out RNA synthesis, capping, and proofreading [58][104]. Remdesivir was originally a drug used for the treatment of hepatitis C and was later shown to be a broad-spectrum antiviral drug with a delayed chain termination mechanism of action [59][60][105,106]. Remdesivir is also being evaluated as an anti-coronavirus drug, and is currently the only drug approved by the US FDA for the treatment of patients with COVID-19. Favipiravir was originally used to treat RNA virus infections such as Ebola and influenza, but a randomized clinical trial found that the drug can bind to the RdRp metal catalytic site of SARS-CoV-2 and produce inhibitory activity [61][107]. Therefore, favipiravir has been urgently approved for the treatment of mild COVID-19 in several countries [62][108]. At present, the drug has entered phase 3 clinical trials for the treatment of COVID-19 in many countries [63][109]. Recent studies have found that suramin, an NNI, can effectively inhibit the activity of SARS-CoV-2 RdRp and prevent the virus from entering cells [64][110]. It has been proved that the anti-COVID-19 mechanism involves two symmetrical suramin molecules binding to RdRp and preventing RNA templates and primers from binding [65][111]. It binds to the active site and prevents nucleotide triphosphates from entering the catalytic site, thereby inhibiting the growth of SARS-CoV-2 [66][112].
2. Research and Development Strategies for Novel Antiviral Drugs
Due to the increasing cost of drug research and development, traditional drug random screening strategies and blind optimization of lead compounds consume a lot of resources and time. In recent years, some new research strategies have been developed, which are listed as follows:
2.1. Nucleic Acid Degradation
Ribonuclease targeting chimera (RIBOTAC) technology converts RNA-binding molecules into RNA-degrading molecules to degrade the viral genome by combining RNA-binding molecules with a small molecule and activating ribonuclease L (RNase L) [67][113]. Haniff et al. (2020) verified through a series of experiments that compound C5 can stabilize the frameshift element and significantly inhibit the frameshift ability of the SARS-CoV-2 frameshift element. At the same time, the structure of compound 39 was modified with the help of RIBOTAC technology, that is, compound 39 was connected to a small molecule that can recruit RNase L through a linking chain of suitable length to achieve the purpose of directional degradation of SARS-CoV-2 mRNA [68][114].
2.2. Protein Degradation
Targeting protein degradation chimera (proteolysis-targeting chimera, PROTAC) molecules can target degradation of proteins [69][115]. PROTAC bifunctional small molecules can bring target proteins and E3 ubiquitin ligases closer together, induce ubiquitination of target proteins and subsequent degradation through the ubiquitin-proteasome pathway [70][116]. Yang et al. (2022) used PROTAC technology to develop anti-HCV molecules, which can induce viral proteasome degradation [71][117]. The PROTAC molecule combined with the ligand of CRL4CRBN can induce HCV NS3/4A protease degradation, proving that protein degradation contributes to its antiviral activity [72][118].
2.3. RNA Interference Application Drugs
RNA interference (RNAi) refers to the specific gene expression silencing mediated by double-stranded RNA [73][119]. ARC-520 is an RNA interference (RNAi)-based drug for the treatment of chronic hepatitis B [74][120]. It can act on HBV covalently closed circular DNA (cccDNA) transcription to degrade mRNA [75][121]. Nonclinical toxicology studies in primates found that ARC-520 may be potentially toxic, and temporarily halted clinical trials of this drug [76][122]. The HBV RNAi drugs currently under development include ARB-1467 (Phase II clinical trial), RG-6004 (Phase I clinical trial), GSK-3389404, and GSK-3228836 (Phase II clinical trial) [77][78][79][123,124,125].
2.4. Capsid Protein Assembly Regulators
The HBV capsid protein assembly regulator can inhibit the replication of the HBV virus by destroying the function of the capsid [80][126]. The HBV capsid can not only protect the viral genome encapsulated in the capsid, but also promote the reverse transcription of pgRNA to form DNA [81][82][127,128]. Heteroaryldihydropyrimidine (HAP) compounds used as HBV capsid assembly modulators (CpAM) can enhance the hydrophobic interaction between adjacent core protein dimers, alter the dimerization angle between the two substances, change their assembly kinetics, and accelerate the degradation of capsid proteins [83][84][129,130]. Electron microscopy results show that HAP compounds can promote the formation of large and irregular particles around which misassembled capsids cannot properly wrap pgRNA, and prevent HBV replication [85][131]. Representative HBV capsid protein assembly regulators include Bay41-4109 (first generation), GIS-4 (second generation, phase 1 clinical trial), and HAP-R10 (third generation, phase I clinical trial) [86][87][132,133].
