Hepatitis E virus (HEV) (family Hepeviridae) is one of the most common human pathogens, causing acute hepatitis and an increasingly recognized etiological agent in chronic hepatitis and extrahepatic manifestations. Not only the members of the species Orthohepevirus A (HEV-A) pathogenic to humans but a genetically highly divergent rat origin hepevirus (HEV-C1) in species Orthohepevirus C (HEV-C) is also able to cause zoonotic infection and symptomatic disease (hepatitis) in humans.
- hepevirus
- orthohepevirus
- rat
- human
- zoonosis
- reservoir
- cross-species transmission
Note:All the information in this draft can be edited by authors. And the entry will be online only after authors edit and submit it.
Definition: Hepatitis E virus (HEV) (family Hepeviridae) is one of the most common human pathogens, causing acute hepatitis and an increasingly recognized etiological agent in chronic hepatitis and extrahepatic manifestations. Recent studies reported that not only are the classical members of the species Orthohepevirus A (HEV-A) pathogenic to humans but a genetically highly divergent rat origin hepevirus (HEV-C1) in species Orthohepevirus C (HEV-C) is also able to cause zoonotic infection and symptomatic disease (hepatitis) in humans.
1. Introduction
Hepatitis E virus (HEV) is one of the most common pathogens causing acute hepatitis in tropical and subtropical areas and an increasingly recognized pathogen in acute, chronic hepatitis and extrahepatic manifestations in developed countries [1]. In general, HEV has a positive-sense, single-stranded 6.6–7.3kb-long RNA genome containing three open reading frames (ORFs) [2]. The ORF1, ORF2, and ORF3 encode the viral non-structural, capsid, and a small immunoreactive proteins, respectively. Hepeviruses are members of the family Hepeviridae and are classified into two genera: Orthohepevirus and Piscihepevirus [2] [2]. Genus Orthohepevirus can be divided into four species: Orthohepevirus A–D. Within the species Orthohepevirus A, at least 8 distinct genotypes (HEV-1 to HEV-8) and several sub-genotypes have been identified [2,3,4][2][3][4]. It should also be noted that there are several further unassigned hepeviruses in the family which predict future significant change in the taxonomy of the virus family. The known sources and transmission routes of HEV infections in human are diverse and—based on the present knowledge—influenced by the genetic variants of the virus. Historically, it is generally thought that HEV is transmitted by human-to-human in developing countries because of the low social and hygienic conditions and only travel-related imported cases occur in developed countries. The most important initial breakthrough was the first detection of HEV in animal species, in swine in 1997 [5], which confirmed, for the first time, the possibility of the animal-to-human transmission of HEV as a zoonosis.
2. Phylogenetic Analysis of Rat Hepeviruses
The members of the species Orthohepevirus C are constituted a distinct group. The nucleotide sequence p-distance within groups of species HEV-C1 and HEV-C2 seems to be low, from 4.2 ± 2.1% to 20.9 ± 6.1% throughout the nucleotide coding region (CDS) of ORF1-ORF4 in each group (data not shown). With a comparison, the mean p-distance scores are 9.7%, 28.9%, 19.7%, 17.4%, 23%, 15.1%, 9.7% within HEV-1, HEV-2, HEV-3, HEV-4, HEV-6, HEV-7, and HEV-8 groups in Orthohepevirus A, respectively. The cluster analysis is a common technique to classify or grouping data (sequences) to provide evolutionary connections or map the relationship between the related species [69,70][6][7]. The rodent hepeviruses form a discrete phylogenetic cluster among species Orthohepevirus C in family Hepeviridae (Figure 31). Based on the phylogenetic analysis (agglomerative, single linkage clustering) these sequences are separable at least four different clusters (C1, C2, “C3”, and “C4”), showing the evolutionary/phylogenetic position of one sequence in among similar/homologous species. Complementing the phylogenetic analysis with k-means cluster analysis, not only the distances between individual sequences can be observed more clearly, but intergroup relationships among rodent origin hepeviruses could be more perceptible (Figure 42) [69][6]. The analysis shows that HEV-C1, C2, and the tentative “C3” group of viruses in Orthohepevirus C could evolve together and may co-evolve with different rodent hosts. The viruses currently thought to be in the HEV-“C4” group are genetically distant from rat hepeviruses, but these viruses are closely related to the hepeviruses found in rodents in family Cricetidae based on the distance calculation between group mean distance: HEV-C1 to C2 is 43.8%, to “C3” is 50.8%, to “C4” is 67.7%, and to unassigned rodent origin hepeviruses is 69.4%, respectively (Figure 42b). Estimating of the average evolutionary divergence over sequence pairs within groups were 24.3% (standard error: 0.54%), 20.4% (s.e.: 0.68%), 8% (s.e.: 0.4%), 11.8% (s.e.: 0.5%), and 65.7% (s.e.: 1.08%) in HEV-C1, HEV-C2, HEV-“C3”, and HEV-“C4” viruses and unassigned rodent-origin (kestrel and common vole) hepeviruses by MEGA software. The mean diversity in the entire population is 51% (s.e.: 0.9%). It seems hepeviruses identified from rodents are genetically much more diverse than hepeviruses phylogenetically classified into cluster Orthohepevirus A. It is conceivable that rodents may be one of the key hosts and/or sources of the spread of hepevirus.
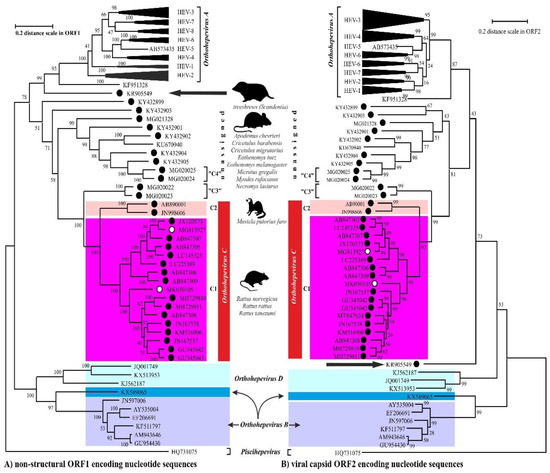
Figure 31. The phylogenetic reconstruction of hepeviruses. Phylogenetic trees are generated based on the (A) ORF1-encoded non-structural polyprotein (left) and (B) ORF2-encoded capsid protein (right) sequences and each tree are rooted to piscihepevirus A (Piscihepevirus) srain Heenan88 (HQ731075). Each phylogenetic tree is focusing on the analysis of the rodent origin HEV-C1 (magenta), HEV-C2 (pink) sequences in Orthohepevirus C (red) complementing with the currently unassigned rodent-borne hepevirus sequences. The putative HEV-“C3” and HEV-“C4” clades in Orthohepevirus C are also highlighted. The rodent hepeviruses are marked with black dot, while rodent origin HEV-C1 in human infections are highlighted by white dot. The reported (potential) source of infection of hepeviruses are shown next to the lineages. The phylogenetic subtrees are compressed based representative genotypes in Orthohepevirus A. The following sequences are used in subtree HEV-1: AY230202, MH918640, AY204877, X98292, L08816, D11092, LC225387, M73218, JF443721, FJ457024; HEV-2: MH809516, KX578717; HEV-3: FJ705359, KP294371, FJ998008, MF959764, LC260517, MF959765, KU513561, MK390971, JQ013794, JQ953664, AB301710, AP003430, KT633715, AF082843, AY115488, AB369689, AB369687, EU723512, EU360977, AB290313, AB248521, JQ026407, AF455784, FJ906895; HEV-4: DQ450072, AB108537, MK410048, AB369688, AB220974, AB074915, KU356182, KF736234, AB197673, AY723745, AJ272108, LC428039, DQ279091; HEV-5: AB573435; HEV-6: AB856243, AB602441; HEV-7: KJ496144, KJ496143; HEV-8: MH410176, KX387867, KX387865. The evolutionary analyses were conducted in MEGA X [71][8], and phylogenetic trees were inferred by using the Maximum Likelihood method and Jukes-Cantor model [72][9]. The tree is drawn to scale, with branch lengths measured in the number of substitutions per site.
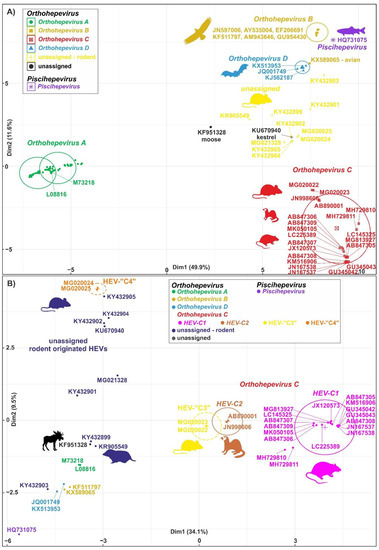
Figure 42. Cluster analysis of hepeviruses based on the complete nucleotide sequences of open reading Figure 13 (ORF1). (A) Plot A represents the distribution of representative hepevirus species in Hepeviridae, and (B) Plot B focuses on the distribution of rodent origin hepeviruses. Piscihepevirus A (purple), Orthohepevirus A (green), B (gold), C (red), and D (blue) and currently unassigned (yellow and black) species in Plot A and Plot B, respectively, are K-means clustered together [73][10]. The rodent origin hepevirus clusters in Orthohepevirus C, HEV-C1 (magenta), HEV-C2 (brown), and two novel clusters, HEV-“C3” (yellow) and HEV-“C4” (orange), are shown in Plot B. Briefly, a total of 101 and 40 complete coding sequence (CDS) of ORF1 sequences in Plot A and Plot B, respectively, are aligned by MEGA using codon based ClustalW alignment method and analyzed by R using seqinr, msa, cluster, factoextra, and randomColoR packages. An identity matrix of pairwise distances from aligned nucleotide sequences was calculated. The matrix scaled and the optimal number of clusters and the means (centres) of each cluster were estimated. The R algorithm code could be provided on request.
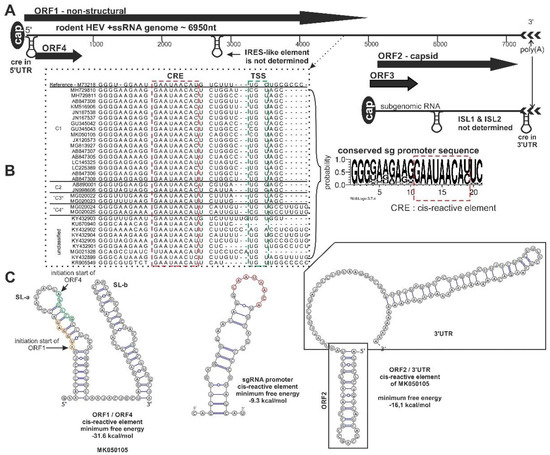
Figure 3. Schematic genome organization of rodent origin hepeviruses in species Orthohepevirus C and closely related, unassigned rodent hepeviruses. (A) The genome representation, (B) the significant nucleotide signals, and (C) the prediction of secondary RNA structural elements in rodent hepeviruses are presented. The alignment of the subgenomic RNA promoter sequence region (black dotted line), including the cis-reactive site (CRE) (red dotted line) and the transcription initiation site (TSS) (green dotted line), are highlighted. The consensus nucleotide Logo is generated from the alignment [11]. The internal ribosome entry site (IRES)-like, internal stem-loop (ISL)1/ISL2, CRE secondary RNA structural elements were predicted by ViennaRNA Web Service using the RNAfold server [12]. Open reading frame (ORF)4 is not present in all members of the orthohepevirus C.
3. Rat Hepevirus (HEV-C1) Infections in Human and Roles in Disease
The zoonotic potential of HEV-C was first suggested, as enzyme-linked immunosorbent assays (ELISAs) with E. coli expressed capsid protein indicated possible subclinical infection with HEV-C among forestry workers in Germany [77][13] and febrile inpatients in Vietnam [78][14]. The first confirmed human infection associated with rat hepevirus was detected from 1 (1.9%) of the 52 solid organ transplanted and immunocompromised patients with persistent hepatitis in Hong Kong, in 2017 [12] [15] (Table 31). The 56-year-old man had abnormal liver function tests at day 59 post-liver transplantation and only the RT-PCRs using the pan-orthohepevirus primers and sequencing confirmed the presence of rat origin HEV-C1 in plasma, feces, saliva, and liver tissue specimens [12][15]. The patient was successfully treated with a 7-month-long oral ribavirin therapy. The source of the infection is potentially associated with rodent droppings in his living environment [12][15]. Based on this result, a 31-month-long prospective investigation found another 6 (0.27%) of 2201 patients with hepatitis and 1 (0.15%) of 659 immunocompromised persons with rat HEV-C1 infections including five patients in two neighbouring districts in Hong Kong between 2017 and 2019 [34] [16] (Table 31). Clinically, the orthohepevirus C hepatitis was generally milder than orthohepevirus A hepatitis with lesser mean peak ALT (alanine-aminotransferase) and bilirubin [34][16]. Expanding the geographic distribution of severe hepatitis caused by rat HEV-C1 was reported from Canada as a potential imported infection from Africa [13][17]. On the other hand, rat HEV-C1 RNA was not detected by RT-PCR from sera collected from 162 patients with acute hepatitis tested positive by HEV IgM ELISA based on viruses in species Orthohepevirus A in Hungary [79][18]. These first reports support that viruses in species Orthohepevirus C are capable of causing human infection and disease, including acute and persistent hepatitis and subclinical infection. These results show that, maybe, immunocompromised patients seem to be more susceptible and at risk for these types of infection than immunocompetent patients. One of the most important questions is the frequency and the transmission mode of the infection because the exact transmission route of orthohepevirus C to human is not confirmed. Humans may have been infected directly via contact with environmental surfaces contaminated by rat droppings [12][15] or indirectly by contamination of water or food products (e.g., consumption of rat meat or adulterated meat products). The transmission of orthohepevirus C from rat (or rodents) through the intermediate animal host (e.g., domestic animals, etc.) to humans is also an open question, as well as the role as blood-borne pathogen through blood transfusion and/or organ transplantation. Mapping the transmission routes of orthohepevirus C are necessary to prevent human infections. Ribavirin could be an effective treatment against HEV-C1 [12][15].
Table 1. Summary of human infections (N = 9) associated with rat origin HEV-C1 virus (species Orthohepevirus C) in the literature. HBV = hepatitis B virus.
| Patient | Underlying Disease | Acute Disease | Potential Source of the Infection | Geographic Region (Year) | GenBank Acc. No | Reference |
|---|---|---|---|---|---|---|
| 56-year-old man | liver transplantation, HBV carrier, immunocompromised | persistent hepatitis | rodent droppings | Hong Kong (2017) | MG813927 | [15] |
| 71-year-old female | rheumatoid arthritis, immunocompromised | acute hepatitis | unknown | Hong Kong (2017) | MN450851 | [16] |
| 67-year-old man | kidney transplantation, immunocompromised | persistent hepatitis | unknown | Hong Kong (2018) | MN450852 | [16] |
| 74-year-old man | kidney transplantation, HBV carrier, immunocompromised | persistent hepatitis | unknown | Hong Kong (2018) | MN450853 | [16] |
| 81-year-old man | prostate cancer | acute hepatitis | unknown | Hong Kong (2019) | MN450856 | [16] |
| 73-year-old man | none, immunocompetent | acute hepatitis | unknown | Hong Kong (2019) | MN450857 | [16] |
| 67-year-old man | metastatic cancer | subclinical | unknown | Hong Kong (?) | MN450858 | [16] |
| 43-year-old man | HIV infection | persistent hepatitis | unknown | Hong Kong (2019) | MN450854 | [16] |
| 49-year-old man | none, immunocompetent | severe acute hepatitis | unknown | Canada(may be imported from Uganda, Africa) (2019) | MK050105 | [17] |
4. Summary and Future Directions
The history of the science of HEVs continues to abound in surprise. It ran a great career from a neglected tropical acute waterborne disease, then a zoonotic infection from animals to humans in developed countries through as an etiological agent in chronic hepatitis and extrahepatic diseases in human. Now, we should re-evaluate the knowledge of hepevirus again. Evidence shows that the animal reservoir of the human hepevirus infections is proven to have been supplemented with rats, and, not only orthohepevirus A, but the member of the orthohepevirus C viruses are also able to cause human zoonosis among both immunocompromised and immunocompetent patients with hepatitis. This novel knowledge has a consequence to the epidemiology, clinical aspects, laboratory diagnosis and prevention of HEV infection in human.
Opening of this novel chapter could not answer yet for several important questions. Further studies needed to investigate how frequent is the HEV-C1 infection in human and what percentage of human HEV disease misdiagnosed because of this hepevirus variant. In addition to this, it should explore the wideness of genetic (and antigen) diversity of human pathogen HEV-C1 in rat, rodents and potential in other animal hosts. It should be studied that are there any differences in zoonotic potential between different rat hepeviruses to human. One of the central issues is to localize the exact hot-spots, the transmission mode(s) from animal to human, and geographic distribution of HEV-C infections. All potential transmission modes should be investigated including the organ and blood transfusion risks in a certain geographic area especially in the situation if the donor is infected asymptomatically. As in orthohepevirus A viruses the full disease spectrum (acute and chronic hepatitis and extrahepatic diseases) of HEV-C1 infections should be investigated in humans. Clinicians are aware that rats are a potential reservoir of human hepevirus infections, too.
HEV-C1 (species Orthohepevirus C) is genetically and antigenically different from viruses in species Orthohepevirus A. At the same time, the presently used human (serological and molecular) diagnostics are designed based on certain genotypes (most of the HEV-1 and HEV-3) of orthohepevirus A viruses. These means that these methods have low sensitivity for HEV-C1 [12,80][15][19]. HEV-C1 infections presently undiagnosed because of i) low antigen cross-immunity in serological test and ii) amplification failure in RT-PCRs because of primer mismatch [12][15]. Serologic tests should be developed which can detect both orthohepevirus A and orthohepevirus C antibodies (at the same time) in humans by ELISA, and we need to extend the coverage of the diagnostic primers and probes for both HEV-A and HEV-C (particularly HEV-C1) in molecular methods.
Confirmation of HEV-C1 infections and symptomatic diseases in humans re-opened the long-lasting and full of surprises story of HEV in human. Rat HEV-C1 is also proving that further studies are needed to explore the full animal reservoir, cross-species transmission, and genetic diversity of hepeviruses potentially threatening humans.
References
- Webb, G.W.; Dalton, H.R. Hepatitis E: An underestimated emerging threat. Ther. Adv. Infect. Dis. 2019, 6, 1–18.
- Meng, X.J.; Anderson, D.A.; Arankalle, V.A.; Emerson, S.U.; Harrison, T.J.; Jameel, S.; Okamoto, H. Hepeviridae; Virus Taxonomy 9th Report of the ICTV; Elsevier Academic Press: London, UK, 2012; pp. 1021–1028.
- Primadharsini, P.P.; Nagashima, S.; Okamoto, H. Genetic variability and evolution of hepatitis E virus. Viruses 2019, 11, 456.
- Smith, D.B.; Izopet, J.; Nicot, F.; Simmonds, P.; Jameel, S.; Meng, X.J.; Norder, H.; Okamoto, H.; van der Poel, W.; Reuter, G.; et al. Update: Proposed reference sequences for subtypes of hepatitis E virus (species Orthohepevirus A). J. Gen. Virol. 2020, 101, 692–698.
- Meng, X.J.; Purcell, R.H.; Halbur, P.G.; Lehman, J.R.; Webb, D.M.; Tsareva, T.S.; Haynes, J.S.; Thacker, B.J.; Emerson, S.U. A novel virus in swine is closely related to the human hepatitis E virus. Proc. Natl. Acad. Sci. USA 1997, 94, 9860–9865.
- James, B.T.; Luczak, B.B.; Girgis, H.Z. MeShClust: An intelligent tool for clustering DNA sequences. Nucl. Acids Res. 2018, 46, e83.
- Balaban, M.; Moshiri, N.; Mai, U.; Jia, X.; Mirarab, S. TreeCluster: Clustering biological sequences using phylogenetic trees. PLoS ONE 2019, 14, e0221068.
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549.
- Jukes, T.H.; Cantor, C.R. Evolution of protein molecules. In Mammalian Protein Metabolism; Munro, H.N., Ed.; Academic Press: New York, NY, USA, 1969; pp. 21–132.
- MacQueen, J. Some methods for classification and analysis of multivariate observations. In Proceedings of the Fifth Berkeley Symposium on Mathematical Statistics and Probability; Le Cam, L.M., Neyman, J., Eds.; University of California Press: Berkeley, CA, USA, 1967; Volume 1, pp. 281–297.
- Crooks, G.E.; Hon, G.; Chandonia, J.M.; Brenner, S.E. WebLogo: A sequence logo generator. Genome Res. 2004, 14, 1188–1190.
- Lorenz, R.; Bernhart, S.H.; Siederdissen, C.H.; Tafer, H.; Flamm, C.; Stadler, P.F.; Hofacker, I.L. ViennaRNA Package 2.0. Algorithms Mol. Biol. 2011, 6, 26.
- Dremsek, P.; Wenzel, J.J.; Johne, R.; Ziller, M.; Hofmann, J.; Groschup, M.H.; Werdermann, S.; Mohn, U.; Dorn, S.; Motz, M.; et al. Seroprevalence study in forestry workers from Eastern Germany using novel genotype 3- and rat hepatitis E virus-specific immunoglobulin G ELISAs. Med. Microbiol. Immunol. 2012, 201, 189–200.
- Shimizu, K.; Hamaguchi, S.; Ngo, C.C.; Li, T.C.; Ando, S.; Yoshimatsu, K.; Yasuda, S.P.; Koma, T.; Isozumi, R.; Tsuda, Y.; et al. Serological evidence of infection with rodent-borne hepatitis E virus HEV-C1 or antigenically related virus in humans. J. Vet. Med. Sci. 2016, 78, 1677–1681.
- Sridhar, S.; Yip, C.C.Y.; Wu, S.; Cai, J.; Zhang, J.X.; Leung, K.H.; Chung, T.W.H.; Chan, F.W.; Chan, W.M.; Teng, J.L.L.; et al. Rat hepatitis E virus as cause of persistent hepatitis after liver transplant. Emerg. Infect. Dis. 2018, 24, 2241–2250.
- Sridhar, S.; Yip, C.C.Y.; Wu, S.; Chew, N.F.S.; Leung, K.H.; Chan, J.F.W.; Zhao, P.S.; Chan, W.M.; Poon, R.W.S.; Tsoi, H.W.; et al. Transmission of rat hepatitis E virus infection to humans in Hong Kong: A clinical and epidemiological analysis. Hepatology 2020.
- Andonov, A.; Robbins, M.; Borlang, J.; Cao, J.; Hatchette, T.; Stueck, A.; Deschambault, Y.; Murnaghan, K.; Varga, J.; Johnston, L. Rat hepatitis E virus linked to severe acute hepatitis in an immunocompetent patient. J. Infect. Dis. 2019, 220, 951–955.
- Pankovics, P.; Némethy, O.; Boros, Á.; Pár, G.; Szakály, P.; Reuter, G. Four-year-long (2014–2017) clinical and laboratory surveillance of hepatitis E virus infections using combined antibody, molecular, antigen and avidity detection methods: Increasing incidence and chronic HEV case in Hungary. J. Clin. Virol. 2020, 124, 104284.
- Sanford, B.J.; Opriessnig, T.; Kenney, S.P.; Dryman, B.A.; Córdoba, L.; Meng, X.J. Assessment of the cross-protective capability of recombinant capsid proteins derived from pig, rat, and avian hepatitis E viruses (HEV) against challenge with a genotype 3 HEV in pigs. Vaccine 2012, 30, 6249–6255.
