Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Małgorzata Natonek - Wiśniewska.
The pig, one of the most important livestock species, is a meaningful source of global meat production. It is necessary to prove whether a food product that a discerning customer selects in a store is actually made from pork or venison, or does not contain it at all. The problem of food authenticity is widespread worldwide, and cases of meat adulteration have accelerated the development of food and the identification methods of feed species. It is worth noting that several different molecular biology techniques can identify a porcine component.
- pig
- wild boar
- species identification
- genetic markers
1. Introduction
The pig is believed to be one of the most important livestock species for humans. It is an excellent animal model for studying the molecular background of several human diseases [1], and above all, it is a meaningful source of global meat production. Pork is widely used in the food industry, but one problem is determining the food’s authenticity, especially in terms of the species. Meat adulteration involves replacing or partially replacing high-value meat with cheaper quality meat [2], which involves economic, quality, safety, and socio-religious issues [3]. Together with pork, products may introduce veterinary drugs banned from the food chain, such as ractopamine, which is forbidden in many countries [4,5][4][5]. Moreover, undeclared pork may be contaminated with harmful microorganisms, such as the endoparasites Toxoplasma and Trichinella [6]. The presence of pork in food may also conflict with religious and cultural practices. Thus, the fraud of undeclared pork in food can affect consumer trust in the meat industry. Because of such illegal food practices, many governments have enacted laws prohibiting similar fraud. To ensure existing regulations are followed, laboratories worldwide have designed and developed various methods to identify animal species in food products, including methods to detect porcine components. According to scientific reports, efforts have also been made to provide a cost-efficient diagnostic tool for distinguishing wild boar (Sus scrofa), domestic pig (Sus scrofa domestica), and their hybrids (Figure 1). This is not easy, as the pig and the wild boar are evolutionarily closely related [7], and thus share many genetic markers. However, the identification methods have changed over time, accompanied by the development of innovative techniques that are now based on DNA testing.
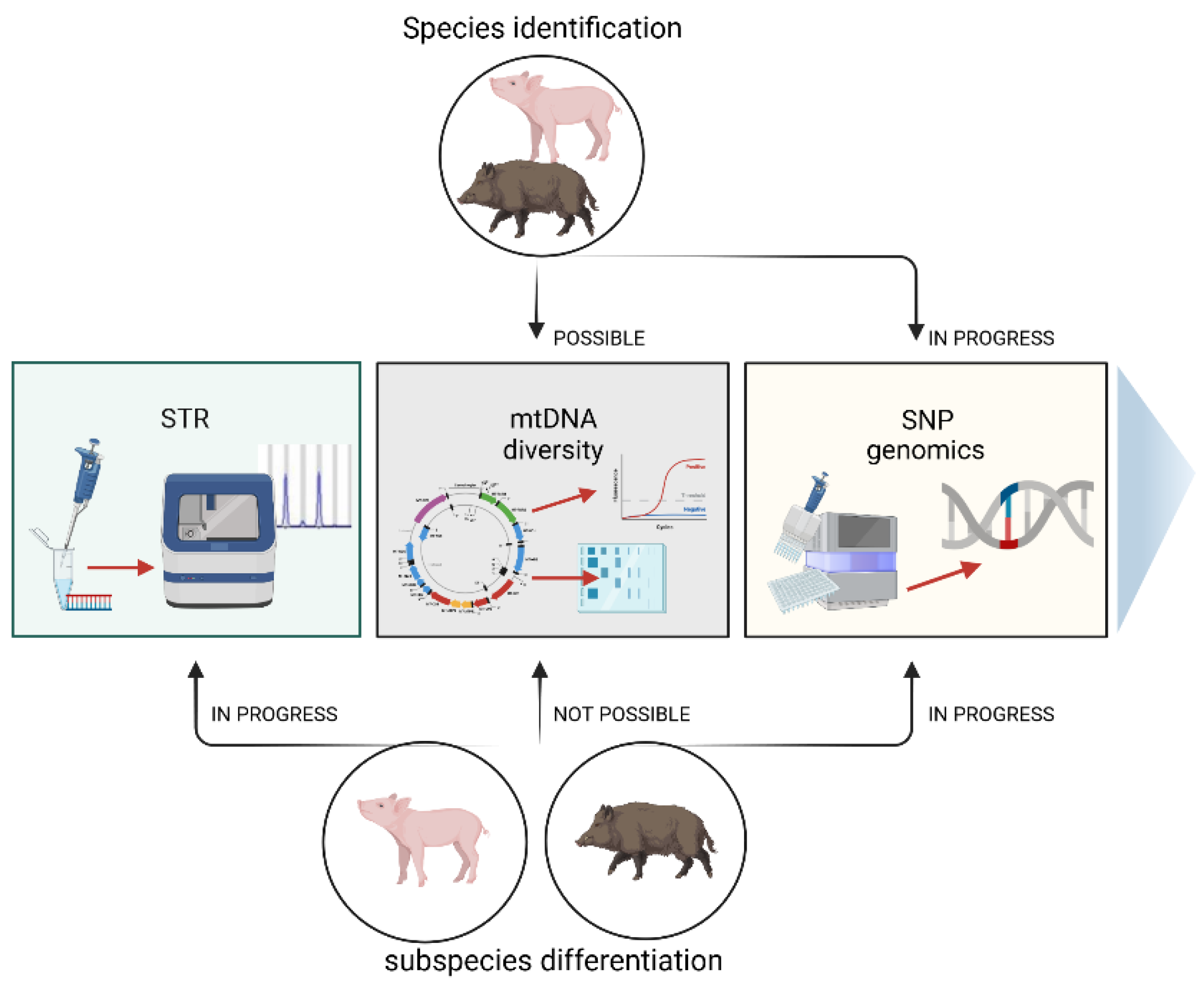
Figure 1. Methods of species identification of domestic pigs and wild boars and distinguishing between both subspecies.
Possibilities of species identification of pigs and wild boars (top of the figure) and distinguishing between both subspecies (bottom of the figure) using different molecular techniques.
2. Pig and Wild Boar Phylogenetics
A considerable number of animal breeds on Earth originated from diverse wild ancestors which, after being domesticated, exhibit a large phenotypic variety [8]. In the study by Groenen et al. [7], a comprehensive analysis of the phylogenetic background of the subspecies structure of wild boars and pigs was provided. The genomes of wild boars and domestic pigs from Europe and Asia revealed separate European and Asian lineages. The study also gave an insight into the history of population size changes. Both lines of wild boars diverged about 1.6–0.8 Myr (million years) ago, resulting in alleles divergence in the millions of loci [7]. Wild boars are the ancestors of modern domestic pigs, and it is believed that multiple domestication events occurred separately for species in Asia and Europe [7,9][7][9].
After domestication, many incidents of the admixture of domestic populations with local wild boars occurred because both species are found in the same habitat [9]. Moreover, there are farms (e.g., in Bulgaria and Sardinia) where pigs are kept in semi-wild conditions. It is highly likely that domestic pigs sporadically crossed with wild boars [10]. The frequent hybridisation of these two breeds was found in the studies on the European population [10,11,12,13][10][11][12][13]. Studies of the MC1R (melanocortin 1 receptor) gene in the Polish population of wild boars also proved the existence of admixed genotypes [14,15][14][15]. Crossbreeding domestic pigs with wild boars is documented and has served hunting purposes, better meat production, better taste, and reduced aggressive behaviour [11,12][11][12]. However, crossbreds have a high risk of pathogen transmission (e.g., ASF, African Swine Fever), which is the primary reason crossbreeding farms are under strict legislation in the European Union [12]. It is highly likely that some wild boars farmed in Poland were released or escaped from the farms and became a potential source of genome admixture in the wild population. Other studies have also suggested such a scenario [11,12,14][11][12][14]. The domestication of the pig and the mixing of domestic pig and wild boar populations cannot be ignored, as they provide the background to the problem of distinguishing between these subspecies.
3. Authentication of Meat from Domestic Pig
The authentication of domestic pig meat is mainly based on mitochondrial DNA (mtDNA). The well-known sequences of the mitochondrial genome allow comparison of the porcine mtDNA with the genome of any species. The advantages of mitochondrial DNA over genomic DNA are due to its properties. The mitochondrial genome is well understood and described in many databases (GenBank, EST, GSS). It is also species-specific and is present in many copies in every organism cell. Moreover, mtDNA is resistant to unfavourable factors, such as high temperature or pressure. This last feature is essential because it allows the identification of DNA in preserved meat products, the production of which involves thermal processing. Properties of mtDNA translate into biological specificity and high sensitivity of pork identification methods, which is very important when investigating highly processed products such as sausages, canned pork, pork fat, or gelatine. However, to date, it is impossible to distinguish pork from venison based on mtDNA variability only.
The heat treatment of food causes fragmentation of the DNA. This may reduce the chance of extracting good quality DNA sufficient for further analysis. In the literature, wresearchers can find many reports on the influence of individual types of heat treatment on DNA quality. Traditional cooking, heating in a microwave [16], or processing pork for canned meal influence the amount of DNA obtained, and the isolate rate remains at a reasonable level. In contrast, DNA extracts from gelatine or meat meal had much worse properties. In the first case, the quantity and quality of DNA described by the absorbance ratio had the values of 400–600 ng/µL and 1.7–1.9, respectively. However, only 15–145 ng/µL were obtained for gelatine or meat meal, with a purity often below 1.6 or above 1.9. These are the difficulties any laboratory faces in food product species identification.
The obtained DNA can be analyzed by many methods. Most of them are based on the PCR reaction, which can be used in several variants such as classical PCR, real-time PCR, sequencing, DNA barcoding and NGS (next-generation sequencing). The advantages and limitations of these methods are presented in Table 1.
Table 1. Advantages and limitations of different methods used for Sus scrofa meat identification.
| Technique/Method | Advantages | Limitations and Difficulties | References | |||
|---|---|---|---|---|---|---|
| PCR | Simple to develop and easy to conduct Can detect small amounts of DNA High specificity species identification Analysis of monoplexes or multiplexes depending on needs Low detection limit Ability to detect processed and heat-treated samples |
No quantitation Lower specificity when amplicon length is short More difficult PCR optimisation in case of multiplex PCR Unequal amplification efficiency which causes variable sensitivity Contaminants could produce false-negative results Lower DNA yield for heat-treated samples |
[17,18,19,20,21] | [17][18][19][20][21] | ||
| PCR RAPD | Detects multi-species, and no previous knowledge of DNA is required Can detect a high level of polymorphism (for example, between similar species) Costs per assay are low |
Unable to differentiate species in mixtures Mistakes during the analysis of degraded or autoclaved materials Difficulties in obtaining reproducible results |
[22,23,24] | [22][23][24] | ||
| PCR-RFLP | Species-specific restriction pattern | The results can be challenging to interpret in the case of entirely unknown species components of the studied object Inadequate for highly processed or meat mixtures |
[25, | [25 | 26] | ][26] |
| Real-Time PCR (quantification and qualification) | Possible quantitative result Very high sensitivity High sample throughput |
Probe-based methods are expensive and time-intensive Dye-based methods are less accurate Quantification PCR requires appropriate reference material Precision of measure is different for different product types qPCR instruments are very costly HRM analysis requires HRM-capable real-time PCR machines and specialised software algorithms |
[27,28,29] | [27][28][29] | ||
| Digital PCR | Better detection limit and accuracy | Little sample volume per reaction Small dynamic range if the number of partitions is limited The risk of falsely low quantification due to molecular dropout Restricted multiplex detection |
[30,31] | [30][31] | ||
| LAMP | Rapid High specificity High amplification efficiency No need for thermal cycling Low operational cost |
Less versatile Little to no multiplexing Less sensitive to inhibitors than PCR in case of complex samples. The method is more complicated than classical PCR owing to the presence of more primers |
[32,33,34,35] | [32][33][34][35] | ||
| DNA barcoding and NGS | High sensitivity and specificity Single-step DNA sequencing and quantification by NGS High throughput detection |
High-quality DNA necessary Long analysis time from DNA extraction to the finish of bioinformatics analysis Complex library preparation protocol for NGS Trace quantity of foreign DNA may cause inaccurate estimation of species Costly equipment to analyse and very highly skilled personnel |
[36,37] | [36][37] |
4. Genetic Markers Used for Pig and Wild-Boar Distinguishing
Molecular techniques to identify animal species components in human food and animal feed use different genetic markers based on genomic or mitochondrial DNA. There are cost-effective and easily performed tests for DNA identification of cattle, horse, and poultry in food and animal feed [17,59][17][38]. Some of the methods used for pigs were discussed above. A reliable method for the differentiation of both subspecies would impact biodiversity, food fraud cases, detection of illegal hunting procedures, and zoonosis prevention. However, whether genomic DNA markers can be useful for analysis performed on DNA extracted from highly processed food products remains a subject of study.
4.1. Mitochondrial DNA Genome Loci
Mitochondrial sequences have been widely studied in the context of phylogenetics in many species [60,61,62][39][40][41], but using mtDNA only for distinguishing wild boar from domestic pig is unreliable. A different approach is needed. Fajardo et al. [63][42] conducted studies on wild boars and commercial pig breeds differentiation based on combining both nucleus (melanocortin 1 receptor, MC1R) and mitochondrial DNA (D-loop) analysis. The analysis of the MC1R gene proved to be more effective for species identification based on the polymorphism of mitochondrial DNA [63][42].
4.2. Melanocortin 1 Receptor Gene (MC1R)
The animal’s coat colour is a visual feature that reflects changes in environment and breeding selection. It is a trait that phenotypically differentiates pigs and wild boars. Wild animal forms are characterised by a dark fur colour, while domestic species have pale coat colours [61][40]. Several genes have been identified as associated with skin and hair pigmentation. MC1R, along with the ASIP gene, regulates the synthesis of two dyes: eumelanin (brown to black) and pheomelanin (crema to red) in melanocytes [64][43]. Both genes express an epistatic interaction [65,66][44][45]. An MC1R allele (E + allele, also called “wild” allele) unique to the wild boar’s population was identified by Kijas et al. [67][46]. Most likely, owing to selection during the breeding work, individuals with the “wild” allele were lost. Implementing PCR-RFLP and real-time PCR protocols to identify four polymorphic MC1R loci gave inconsistent results. In Greece, a population of pigs and wild boars was distinguished using this marker [68][47]. However, further studies revealed that the “wild” allele was also present in some domestic breeds, and “domestic” alleles were found in wild boar samples [11,14,69][11][14][48].
4.3. Nuclear Receptor Subfamily 6, Group A, Member 1 (NR6A1)
Owing to increasing meat efficiency, domestic pig breeds are characterised by more vertebrae. This is another trait that differentiates wild boars and European commercial pigs. In the wild boar, the number of vertebrae is 19 and in pig breeds it is 21–23. A proline to leucine substitution at codon 192 (p.Pro192Leu) in the nuclear receptor subfamily 6, group A, member 1 (NR6A1) gene was shown to be the most likely causative mutation underlying the QTL (Quantitative Trait Loci) [70][49].
Since neither MC1R nor NR6A1 studies as single markers proved to be an effective tool to differentiate both species, the combination of both genes was studied. In general, the combination of single nucleotide polymorphisms in MC1R and NR6A1 genes was tested using real-time PCR [11,71][11][50] or multiplexed using the SNaPshot Multiplex System method [72][51]. The results were promising.
4.4. Short Tandem Repeats (STRs)
Short tandem repeats (STRs, also known as microsatellites) are genetic markers that are commonly used in parentage testing and verification [73][52]. These markers are specific for the species, so they seemed good candidates to distinguish wild boar and pig. The hybridisation between pigs and wild boars based on STRs and MC1R was studied by Nikolov et al. [74][53]. STRs combined with SNPs in MC1R, NR6A1 genes or mitochondrial D-loop have also been analyzed [10,75][10][54].
Bayesian clustering based on 10 STRs and mitochondrial D-loop polymorphic loci indicates that the wild and domestic forms are not divergent. Wild boars and domestic pigs share the most common mtDNA haplotypes, and microsatellite alleles weakly resolve both groups when examining a phylogenetic tree [10]. Nikolov et al. [74][53] proved the introgression of wild boar to Balkanian domestic pig breeds by studying MC1R genotypes and 10 STRs. In the population from Balkanste, the introgression in wild boar is more apparent than the introgression in domestic pigs from Western Europe [74][53]. Lorenzini et al. [75][54] implemented the STRs protocol combined with MC1R and NR6A1 to identify both subspecies for forensic purposes. In this case, STRs were introduced to group individuals into the parental population. However, this marker type does not efficiently identify recent hybrids. STRs alone identify lower hybrids than genotyping polymorphic loci, along with MC1R and NR6A1 [75][54].
References
- Bassols, A.; Costa, C.; Eckersall, P.D.; Osada, J.; Sabrià, J.; Tibau, J. The pig as an animal model for human pathologies: A proteomics perspective. Proteom.—Clin. Appl. 2014, 8, 715–731.
- Ropodi, A.; Pavlidis, D.; Mohareb, F.; Panagou, E.; Nychas, G.-J. Multispectral image analysis approach to detect adulteration of beef and pork in raw meats. Food Res. Int. 2014, 67, 12–18.
- Alamprese, C.; Casale, M.; Sinelli, N.; Lanteri, S.; Casiraghi, E. Detection of minced beef adulteration with turkey meat by UV–vis, NIR and MIR spectroscopy. LWT 2013, 53, 225–232.
- Cai, S.; Kong, F.; Xu, S. Detection of porcine-derived ingredients from adulterated meat based on real-time loop-mediated isothermal amplification. Mol. Cell Probes 2020, 53, 101609.
- Niño, A.M.; Granja, R.H.; Wanschel, A.C.; Salerno, A.G. The challenges of ractopamine use in meat production for export to European Union and Russia. Food Control 2017, 72, 289–292.
- King, H.; Bedale, W. Hazard Analysis and Risk-Based Preventive Controls: Improving Food Safety in Human Food Manufacturing for Food Businesses; Academic Press: Cambridge, MA, USA, 2017; ISBN 978-0-12-809475-4.
- Groenen, M.A.M.; Archibald, A.L.; Uenishi, H.; Tuggle, C.K.; Takeuchi, Y.; Rothschild, M.F.; Rogel-Gaillard, C.; Park, C.; Milan, D.; Megens, H.-J.; et al. Analyses of pig genomes provide insight into porcine demography and evolution. Nature 2012, 491, 393–398.
- Yang, B.; Cui, L.; Perez-Enciso, M.; Traspov, A.; Crooijmans, R.P.M.A.; Zinovieva, N.; Schook, L.B.; Archibald, A.; Gatphayak, K.; Knorr, C.; et al. Genome-wide SNP data unveils the globalization of domesticated pigs. Genet. Sel. Evol. 2017, 49, 1–15.
- Larson, G.; Burger, J. A population genetics view of animal domestication. Trends Genet. 2013, 29, 197–205.
- Scandura, M.; Iacolina, L.; Crestanello, B.; Pecchioli, E.; DI Benedetto, M.F.; Russo, V.; Davoli, R.; Apollonio, M.; Bertorelle, G. Ancient vs. recent processes as factors shaping the genetic variation of the European wild boar: Are the effects of the last glaciation still detectable? Mol. Ecol. 2008, 17, 1745–1762.
- Fontanesi, L.; Ribani, A.; Scotti, E.; Utzeri, V.; Veličković, N.; Dall’Olio, S. Differentiation of meat from European wild boars and domestic pigs using polymorphisms in the MC1R and NR6A1 genes. Meat Sci. 2014, 98, 781–784.
- Iacolina, L.; Pertoldi, C.; Amills, M.; Kusza, S.; Megens, H.-J.; Bâlteanu, V.A.; Bakan, J.; Cubric-Curik, V.; Oja, R.; Saarma, U.; et al. Hotspots of recent hybridization between pigs and wild boars in Europe. Sci. Rep. 2018, 8, 17372.
- Mary, N.; Iannuccelli, N.; Petit, G.; Bonnet, N.; Pinton, A.; Barasc, H.; Amélie, F.; Calgaro, A.; Grosbois, V.; Servin, B.; et al. Genome-wide analysis of hybridization in wild boar populations reveals adaptive introgression from domestic pig. Evol. Appl. 2022, 15, 1115–1128.
- Dzialuk, A.; Zastempowska, E.; Skórzewski, R.; Twarużek, M.; Grajewski, J. High domestic pig contribution to the local gene pool of free-living European wild boar: A case study in Poland. Mammal Res. 2017, 63, 65–71.
- Babicz, M.; Pastwa, M.; Skrzypczak, E.; Buczyński, J.T. Variability in themelanocortin 1 receptor(MC1R) gene in wild boars and local pig breeds in Poland. Anim. Genet. 2013, 44, 357–358.
- Musto, M. DNA Quality and Integrity of Nuclear and Mitochondrial Sequences from Beef Meat as Affected by Different Cooking Methods. Food Technol. Biotechnol. 2011, 49, 523–528.
- Natonek-Wiśniewska, M.; Krzyścin, P.; Piestrzyńska-Kajtoch, A. The species identification of bovine, porcine, ovine and chicken components in animal meals, feeds and their ingredients, based on COX I analysis and ribosomal DNA sequences. Food Control 2013, 34, 69–78.
- Tan, L.L.; Ahmed, S.A.; Ng, S.K.; Citartan, M.; Raabe, C.A.; Rozhdestvensky, T.S.; Tang, T.H. Rapid detection of porcine DNA in processed food samples using a streamlined DNA extraction method combined with the SYBR Green real-time PCR assay. Food Chem. 2019, 309, 125654.
- Prusakova, O.V.; Glukhova, X.A.; Afanas’Eva, G.V.; Trizna, Y.A.; Nazarova, L.F.; Beletsky, I.P. A simple and sensitive two-tube multiplex PCR assay for simultaneous detection of ten meat species. Meat Sci. 2018, 137, 34–40.
- Natonek-Wiśniewska, M.; Krzyścin, P. Evaluation of the suitability of mitochondrial DNA for species identification of microtraces and forensic traces. Acta Biochim. Pol. 2017, 64, 705–708.
- Man-an, S.K.A.; Fadilah, A.R.; Mardhiyyah, S. Contemporary Issues and Development in the Global Halal Industry; Ab.Manan, S.K., Abd Rahman, F., Sahri, M., Eds.; Springer: Singapore, 2016; ISBN 978-981-10-1450-5.
- Mane, B.G.; Tanwar, V.K.; Girish, P.S.; Sharma, D.; Dixit, V.P. RAPD Markers for Differentiation of Meat Species. Indian J. Vet. Res. 2008, 17, 9–13.
- Nakyinsige, K.; Man, Y.B.C.; Sazili, A.Q. Halal authenticity issues in meat and meat products. Meat Sci. 2012, 91, 207–214.
- Chappal-war, A.M.; Pathak, V.; Goswami, M.; Sharma, B.; Singh, P.; Mishra, R. Recent Novel Techniques Applied for Detection of Meat Adulteration and Fraudulent Practices. Indian J. Vet. Public Health 2020, 7, 1–6.
- Rahmati, S.; Julkapli, N.M.; Yehye, W.A.; Basirun, W.J. Identification of meat origin in food products–A review. Food Control 2016, 68, 379–390.
- Hossain, M.A.M.; Ali, E.; Hamid, S.B.A.; Asing; Mustafa, S.; Desa, M.N.M.; Zaidul, I.S.M. Double Gene Targeting Multiplex Polymerase Chain Reaction–Restriction Fragment Length Polymorphism Assay Discriminates Beef, Buffalo, and Pork Substitution in Frankfurter Products. J. Agric. Food Chem. 2016, 64, 6343–6354.
- Lopez-Oceja, A.; Nuñez, C.; Baeta, M.; Gamarra, D.; de Pancorbo, M. Species identification in meat products: A new screening method based on high resolution melting analysis of cyt b gene. Food Chem. 2017, 237, 701–706.
- Thanakiatkrai, P.; Kitpipit, T. Meat species identification by two direct-triplex real-time PCR assays using low resolution melting. Food Chem. 2017, 233, 144–150.
- Natonek-Wiśniewska, M.; Krzyścin, P. Development of Easy and Effective Real-Time PCR tests to Identify Bovine, Porcine, and Ovine Components in Food. Zywnosc Nauk. Technol. Jakosc/Food Sci. Technol. Qual. 2015, 22, 73–84.
- Shehata, H.R.; Li, J.; Chen, S.; Redda, H.; Cheng, S.; Tabujara, N.; Li, H.; Warriner, K.; Hanner, R. Droplet digital polymerase chain reaction (ddPCR) assays integrated with an internal control for quantification of bovine, porcine, chicken and turkey species in food and feed. PLoS ONE 2017, 12, e0182872.
- Köppel, R.; Ganeshan, A.; Weber, S.; Pietsch, K.; Graf, C.; Hochegger, R.; Griffiths, K.; Burkhardt, S. Duplex digital PCR for the determination of meat proportions of sausages containing meat from chicken, turkey, horse, cow, pig and sheep. Eur. Food Res. Technol. 2019, 245, 853–862.
- Lee, S.-Y.; Kim, M.-J.; Hong, Y.; Kim, H.-Y. Development of a rapid on-site detection method for pork in processed meat products using real-time loop-mediated isothermal amplification. Food Control 2016, 66, 53–61.
- Ran, G.; Ren, L.; Han, X.; Liu, X.; Li, Z.; Pang, D.; Ouyang, H.; Tang, X. Development of a Rapid Method for the Visible Detection of Pork DNA in Halal Products by Loop-Mediated Isothermal Amplification. Food Anal. Methods 2015, 9, 565–570.
- Riztyan; Takasaki, K.; Yamakoshi, Y.; Futo, S. Single-Laboratory Validation of Rapid and Easy DNA Strip for Porcine DNA Detection in Beef Meatballs. J. AOAC Int. 2018, 101, 1653–1656.
- Tasrip, N.A.; Mokhtar Khairil, N.F.; Hanapi, U.K.; Abdul Manaf, Y.N.; Ali, M.E.; Cheah, Y.K. No Title Loop Mediated Isothermal Amplification; a Review on Its Application and Strategy in Animal Species Authentication of Meat Based Food Products. Int. Food Res. J. 2019, 26, 1–10.
- Naaum, A.M.; Shehata, H.R.; Chen, S.; Li, J.; Tabujara, N.; Awmack, D.; Lutze-Wallace, C.; Hanner, R. Complementary molecular methods detect undeclared species in sausage products at retail markets in Canada. Food Control 2018, 84, 339–344.
- Haynes, E.; Jimenez, E.; Pardo, M.A.; Helyar, S.J. The future of NGS (Next Generation Sequencing) analysis in testing food authenticity. Food Control 2019, 101, 134–143.
- Safdar, M.; Junejo, Y.; Arman, K.; Abasıyanık, M. A highly sensitive and specific tetraplex PCR assay for soybean, poultry, horse and pork species identification in sausages: Development and validation. Meat Sci. 2014, 98, 296–300.
- Fang, M.; Andersson, L. Mitochondrial diversity in European and Chinese pigs is consistent with population expansions that occurred prior to domestication. Proc. R. Soc. B Boil. Sci. 2006, 273, 1803–1810.
- Bruford, M.W.; Bradley, D.G.; Luikart, G. DNA markers reveal the complexity of livestock domestication. Nat. Rev. Genet. 2003, 4, 900–910.
- Koseniuk, A.; Słota, E. Mitochondrial control region diversity in Polish sheep breeds. Arch. Anim. Breed. 2016, 59, 227–233.
- Fajardo, V.; González, I.; Martín, I.; Rojas, M.; Hernández, P.E.; Garcia, T.; Martín, R. Differentiation of European wild boar (Sus scrofa scrofa) and domestic swine (Sus scrofa domestica) meats by PCR analysis targeting the mitochondrial D-loop and the nuclear melanocortin receptor 1 (MC1R) genes. Meat Sci. 2008, 78, 314–322.
- Klungland, H.; Våge, D.I. Pigmentary Switces in Domestic Animal Species. Ann. N. Y. Acad. Sci. 2003, 994, 331–338.
- Lu, D.; Willard, D.; Patel, I.R.; Kadwell, S.; Overton, L.; Kost, T.; Luther, M.; Chen, W.; Woychik, R.P.; Wilkison, W.O.; et al. Agouti Protein Isan Antagonist of the Melanocyte-Stimulating-Hormone Receptor. Nature 1994, 371, 799–802.
- Ollmann, M.M.; Lamoreux, M.L.; Wilson, B.D.; Barsh, G.S. Interaction of Agouti protein with the melanocortin 1 receptor in vitro and in vivo. Genes Dev. 1998, 12, 316–330.
- Kijas, J.M.H.; Wales, R.; Törnsten, A.; Chardon, P.; Moller, M.; Andersson, L. Melanocortin Receptor 1 (MC1R) Mutations and Coat Color in Pigs. Genetics 1998, 150, 1177–1185.
- Koutsogiannouli, E.A.; Moutou, K.A.; Sarafidou, T.; Stamatis, C.; Mamuris, Z. Detection of hybrids between wild boars (Sus scrofa scrofa) and domestic pigs (Sus scrofa f. domestica) in Greece, using the PCR-RFLP method on melanocortin-1 receptor (MC1R) mutations. Mamm. Biol. 2010, 75, 69–73.
- Koseniuk, A.; Smołucha, G.; Natonek-Wiśniewska, M.; Radko, A.; Rubiś, D. Differentiating Pigs from Wild Boars Based on NR6A1 and MC1R Gene Polymorphisms. Animals 2021, 11, 2123.
- Mikawa, S.; Morozumi, T.; Shimanuki, S.-I.; Hayashi, T.; Uenishi, H.; Domukai, M.; Okumura, N.; Awata, T. Fine mapping of a swine quantitative trait locus for number of vertebrae and analysis of an orphan nuclear receptor, germ cell nuclear factor (NR6A1). Genome Res. 2007, 17, 586–593.
- Kaltenbrunner, M.; Mayer, W.; Kerkhoff, K.; Epp, R.; Rüggeberg, H.; Hochegger, R.; Cichna-Markl, M. Differentiation between wild boar and domestic pig in food by targeting two gene loci by real-time PCR. Sci. Rep. 2019, 9, 1–11.
- Beugin, M.-P.; Baubet, E.; De Citres, C.D.; Kaerle, C.; Muselet, L.; Klein, F.; Queney, G. A set of 20 multiplexed single nucleotide polymorphism (SNP) markers specifically selected for the identification of the wild boar (Sus scrofa scrofa) and the domestic pig (Sus scrofa domesticus). Conserv. Genet. Resour. 2017, 9, 671–675.
- Radko, A.; Smołucha, G.; Koseniuk, A. Microsatellite DNA Analysis for Diversity Study, Individual Identification and Parentage Control in Pig Breeds in Poland. Genes 2021, 12, 595.
- Nikolov, I.; Stoeckle, B.; Markov, G.; Kuehn, R. Substantial hybridisation between wild boars (Sus scrofa scrofa) and East Balkan pigs (Sus scrofa f. domestica) in natural environment as a result of semi-wild rearing in Bulgaria. Czech J. Anim. Sci. 2017, 62, 1–8.
- Lorenzini, R.; Fanelli, R.; Tancredi, F.; Siclari, A.; Garofalo, L. Matching STR and SNP genotyping to discriminate between wild boar, domestic pigs and their recent hybrids for forensic purposes. Sci. Rep. 2020, 10, 1–10.
More
