The modern cultivated wheat has passed a long evolution involving origin of wild emmer (WEM), development of cultivated emmer, formation of spelt wheat and finally establishment of modern bread wheat and durum wheat. During this evolutionary process, rapid alterations and sporadic changes in wheat genome took place, due to hybridization, polyploidization, domestication, and mutation. This has resulted in some modifications and a high level of gene loss. As a result, the modern cultivated wheat does not contain all genes of their progenitors. These lost genes are novel for modern wheat improvement.
- gene modification
- wild emmer wheat
- evolution and domestication
- novel genes
- trait enhancement
The evolution of wheat went through a long and multiple processes, including natural hybridization, polyploidization, domestication, and mutation that took place for more than 300,000 years, making it be a distinct model plant for evolutionary study [1][2][3]. At the early stage of evolution, it was difficult for new species to survive as a combination of different genome enveloped within one nucleus and followed by chromosome doubling resulted in severe genetic stress [4][5]. To cope up with stress, they had to face several challenges, such as rapid differentiation of homologous chromosomes for preventing inter-genomic pairing or securing intra-genomic pairing at meiosis and arranging inter-genomic genetic expression for harmonic coexistence [6]. These challenges are meet up through immediate genomic changes, including chromosome re-patterning, chromatin re-modelling, and molecular alteration [7]. Additionally, numerous morphological and physiological changes occurred during domestication which termed as ‘domestication syndrome’, including changes in seed dispersal mode, in plant architecture, increase in kernel size, loss of seed dormancy and change in nutrient content [8]. Even some genes get lost forever during evolution. Natural wheat and related allopolyploids have 2–10% less DNA than the sum of their parents which indicates elimination of DNA during evolution [9][10].
1. Evolution of Wheat
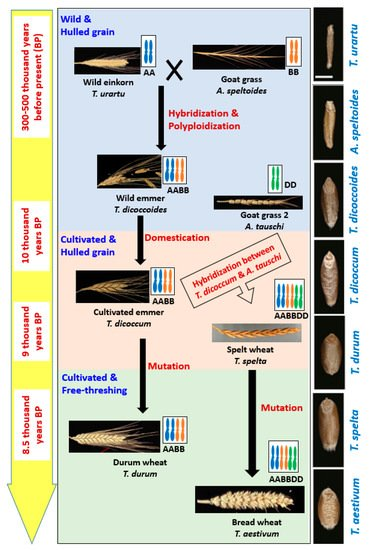
Wheat belongs to the genus Triticum which includes six species: Triticum monococcum (AA); Triticum urartu (AA); Triticum turgidum L. (AABB); Triticum timopheevii (AAGG); Triticum aestivum (AABBDD); and Triticum zhukovskyi (AAAAGG), which can be grouped into three categories: Monococcum (2x), Dicoccoidiea (4x) and Triticum (6x). The reason behind these diversified species is an evolutionary process which is truly a very complex that started at prehistoric Stone Age [11][12][13] (Figure 1).
Figure 1. The central flow chart shows the evolution of wheat along with modification in spike size and spike threshability. Left side yellow colored bar indicates the approximate time of those events happened, and right side black colored bar shows the gradual changes in grain size and shape during evolution.
Wild emmer (WEM, T. dicoccoides) was produced through hybridization between wild einkorn and Goat Grass 1 [11][12][13]. Cultivated emmer (T. turgidum spp. Dicoccum) evolved gradually through subconscious selection from WEM by ancient people, particularly by hunter-gatherers in the Fertile Crescent region. Two ideas were found describing domestication of WEM: (a) Domestication was started in the northern region of the fertile crescent and instant spread to the south or vice-versa, (b) domestication occurred in both northern and southern part independently [14]. However, later, some other archaeological evidence strongly suggested independent domestication and cultivation of WEM in multiple sites . Domesticated emmer hybridized spontaneously later with another wild genotype called Goat Grass 2 and produced hexaploid spelt wheat [15][16]. Both cultivated emmer and spelt wheat were characterized with hulled grain. Free-threshing durum and bread wheat were originated from enclosed cultivated emmer and spelt wheat respectively through natural mutation .
2. Genomic Changes through Domestication
According to the history of wheat evolution, only wild einkorn and WEM wheats went through the early domestication selection . T. monococcum (2x) was domesticated from the wild progenitor species T. boeoticum in the Fertile Crescent and has never been involved in the evolution of hexaploid bread wheat or tetraploid durum wheat. The wild diploid T. urartu (AA) was the progenitor of hexaploid wheat and played an essential role in wheat evolution [17]. Substantial genetic erosion occurred through the domestication process of wheat and that erosion was further reinforced during modern breeding processes [18][19][20]. Consequently, loss of diversity, selective sweeps and adaptive diversification have occurred that caused considerable genetic modification [21]. For example, nucleotide diversity at 21 gene loci was analyzed in wild, domesticated, cultivated durum and bread wheats, and revealed that diversity was reduced in cultivated forms during domestication by 69% in bread wheat and 84% in durum wheat [22].
At the very beginning of the domestication process, the major domestication trait was the seed dispersal mode . Certainly plants with reduced spikelet shattering at maturity had been domesticated, which was considered as a key feature in preventing natural yield losses . In addition to the yield, the other major domestication-related traits includes glume tenacity, spike compactness and flowering time (Table 1).
Table 1: Traits and gene transformed due to domestication
|
Trait |
Transformation due to Domestication |
Gene name |
|
Brittle rachis to non-brittleness |
brittle rachis (Br1, Br2 and Br3) |
|
|
Tough glumes to soft glumes |
tough glumes (Tg1) soft glumes (sog) |
|
|
Spike compactness
|
Non threshing or difficult threshing to free-threshing |
threshability gene (Q gene) |
|
Increase in seed size and weight. Long and thin grains to wider and shorter grain. |
grain size (GS3) grain weight (GW2) seed width (SW5) |
|
|
Flowering time [27] |
Domestication involved selection of spring wheat that lack of vernalization and specific photoperiod requirement. |
An allele on 5A of wild emmer (similar to VRN1 and at collinear position with Ppd) |
3. Genomic Changes through Polyploidization
During the polyploidization of wheat, rapid alteration and several genomic changes occurred in nature. Such phenomena can be divided into two groups: Revolutionary changes, and evolutionary changes . These changes can be of various types, including the elimination of both low-copy and high-copy DNA sequences, intergenomic disruption of DNA sequences, DNA methylation, deletion of rRNA, gene loss, suppression or activation of gene, chromatin modification and remodeling, heterochromatinization, sub-functionalization, and neo-functionalization [28]. Besides, hybridizations that occurred during the evolution of wheat also resulted in some significant genetic changes. For examples, in the crossing product of Aegilops sharonensis and Aegilops umbellulate, 14% loci from Ae. sharonensis and 0.5% loci from Ae. umbellulate were lost [29].
However, it is evident that evolution results in several genomic changes. For example: Nucleolus organizing regions (NORs) is present on different chromosomes (1A, 5A, 1B, 6B, and 5D) of diploid wheat [30]. Its activity is associated with the size of the intergenic regulatory region and the status of cytosine methylation [31][32]. However, NORs from the A genome are largely lost during the evolution of synthetic tetraploid wheat, due to asymmetric transcription and epigenetic modifications during polyploidization. In hybrids, NORs from both parents were expressed. After chromosome doubling, it became silenced in one parent (A genome), due to increased DNA methylation. In stable synthetic tetraploid wheat, rDNA from only the B genome was present. In the case of bread wheat, the rDNA loci form both A and D genomes were largely eliminated during evolution .
Chromosome-specific sequences (CSSs) occur in only one homologous chromosome pair, i.e., 1A and 1A. These types of sequences were present in all diploid species. However, after polyploidization, CSSs from one genome were eliminated immediately or after some generations. As a consequence, in hexaploid or tetraploid, these sequences occur only in one homologous pair but absent from the homeologous chromosome [33]. Meanwhile, allopolyploidization results in rapid non-random deletion of specific non-coding, low-copy and high-copy DNA [34]. Again, sometimes some genes of the A and B genomes get suppressed upon adding of D genome which is known as intergenomic suppression. This is called intergenomic suppression. For example, the rust resistance gene(s) present in the A or B genome was suppressed by a gene present in the long arm of chromosome 7D [35].
3.1. Genomic Changes through Natural Mutation
Through the centuries, natural mutation resulted in significant changes in the genomic structures of wheat, which contributed substantially in the genetic evolutionary process of wheat. In general, mutation generated new alleles, while recombination created novel allele combinations. Accumulation of new mutations in older polyploid species, such as WEM, results in increased diversity and more uniform distribution across the genome [36]. For example, Genetic studies revealed that two recessive alleles at two major loci (Br-A1 and Br-B1) controlling non-brittle rachis raised through mutation during domestication [37]. One of the most important genomic changes is the evolution of free-threshing wheat as a result of several major and minor mutation events. A single major gene Q on chromosome 5AL is responsible for free-threshing of modern bread and durum wheat, whereas the recessive q allele is for non-free-threshing wild wheats . A recent study showed that the Q allele arose from q allele through a gain of function mutation [38]. Free-threshabiliy is also related with tenacious glume (Tg) gene, because Tg inhibits the expression of Q gene. QTL correspond to the Tg gene is located on 2D and 2B chromosome. Free-threshing phenotype evolved when mutation transformed Tg into tg. Therefore free-threshing common bread wheat (QQ5Atgtg2Btgtg2D) and free-threshing durum wheat (QQ5Atgtg2B) have mutant alleles at each of the important threshability loci.
References
- Peng, J.H.; Sun, D.; Nevo, E. Domestication evolution, genetics and genomics in wheat. Mol. Breed. 2011, 28, 281.
- Matsuoka, Y. Evolution of polyploid Triticum wheats under cultivation: The role of domestication, natural hybridization and allopolyploid speciation in their diversification. Plant Cell Physiol. 2011, 52, 750–764.
- Doebley, J.F.; Gaut, B.S.; Smith, B.D. The molecular genetics of crop domestication. Cell 2006, 127, 1309–1321.
- Feldman, M.; Levy, A.A. Genome evolution in allopolyploid wheat—A revolutionary reprogramming followed by gradual changes. J. Genet. Genom. 2009, 36, 511–518.
- Zhao, N.; Xu, L.; Zhu, B.; Li, M.; Zhang, H.; Qi, B.; Xu, C.; Han, F.; Liu, B. Chromosomal and genome-wide molecular changes associated with initial stages of allohexaploidization in wheat can be transit and incidental. Genome 2011, 54, 692–699.
- Feldman, M.; Levy, A. Allopolyploidy–A shaping force in the evolution of wheat genomes. Cytogenet. Genome Res. 2005, 109, 250–258.
- Guo, X.; Han, F. Asymmetric epigenetic modification and elimination of rDNA sequences by polyploidization in wheat. Plant Cell 2014, 26, 4311–4327.
- Peleg, Z.; Fahima, T.; Korol, A.B.; Abbo, S.; Saranga, Y. Genetic analysis of wheat domestication and evolution under domestication. J. Exp. Bot. 2011, 62, 5051–5061.
- Eilam, T.; Anikster, Y.; Millet, E.; Manisterski, J.; Feldman, M. Nuclear DNA amount and genome downsizing in natural and synthetic allopolyploids of the genera Aegilops and Triticum. Genome 2008, 51, 616–627.
- Feldman, M.; Levy, A.A. Genome evolution due to allopolyploidization in wheat. Genetics 2012, 192, 763–774.
- Peng, J.; Sun, D.; Nevo, E. Wild emmer wheat,'Triticum dicoccoides', occupies a pivotal position in wheat domestication process. Aust. J. Crop Sci. 2011, 5, 1127.
- Dvorak, J.; Akhunov, E.D. Tempos of gene locus deletions and duplications and their relationship to recombination rate during diploid and polyploid evolution in the Aegilops-Triticum alliance. Genetics 2005, 171, 323–332.
- Huang, X.; Börner, A.; Röder, M.; Ganal, M. Assessing genetic diversity of wheat (Triticum aestivum L.) germplasm using microsatellite markers. Theor. Appl. Genet. 2002, 105, 699–707.
- Feldman, M.; Kislev, M.E. Domestication of emmer wheat and evolution of free-threshing tetraploid wheat. Isr. J. Plant Sci. 2007, 55, 207–221.
- Dvorak, J.; Luo, M.; Yang, Z. Genetic evidence on the origin of Triticum aestivum L. In The Origins of Agriculture and Crop Domestication, Proceedings of the Harlan Symposium, Aleppo, Syria, 10–14 May 1997; ICARDA: Aleppo, Syria, 1998.
- Matsuoka, Y.; Nasuda, S. Durum wheat as a candidate for the unknown female progenitor of bread wheat: An empirical study with a highly fertile F 1 hybrid with Aegilops tauschii Coss. Theor. Appl. Genet. 2004, 109, 1710–1717.
- Zohary, D.; Hopf, M. Domestication of Plants in the Old World: The Origin and Spread of Cultivated Plants in West Asia; Europe, and the Nile Valley Oxford University Press: New York, NY, USA, 2000.
- Fu, Y.-B.; Somers, D.J. Genome-wide reduction of genetic diversity in wheat breeding. Crop Sci. 2009, 49, 161–168.
- Nevo, E. Ecological genomics of natural plant populations: The Israeli perspective. In Plant Genomics; Springer: Berlin, Germany, 2009; pp. 321–344.
- Nevo, E. Triticum. In Wild Crop Relatives: Genomic and Breeding Resources, Springer: Berlin, Germany, 2011; pp. 407–456.
- Brown, A.H. Variation under domestication in plants: 1859 and today. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2010, 365, 2523–2530.
- Haudry, A.; Cenci, A.; Ravel, C.; Bataillon, T.; Brunel, D.; Poncet, C.; Hochu, I.; Poirier, S.; Santoni, S.; Glémin, S. Grinding up wheat: A massive loss of nucleotide diversity since domestication. Mol. Biol. Evol. 2007, 24, 1506–1517.
- Gill, B.S.; Friebe, B.; Raupp, W.J.; Wilson, D.L.; Cox, T.S.; Sears, R.G.; Brown‐Guedira, G.L.; Fritz, A.K. Wheat genetics resource center: The first 25 years. Adv. Agron. 2006, 89, 73–136.
- Nalam, V.J.; Vales, M.I.; Watson, C.J.; Johnson, E.B.; Riera-Lizarazu, O. Map-based analysis of genetic loci on chromosome 2D that affect glume tenacity and threshability, components of the free-threshing habit in common wheat (Triticum aestivum L.). Theor. Appl. Genet. 2007, 116, 135–145.
- Jantasuriyarat, C.; Vales, M.; Watson, C.; Riera-Lizarazu, O. Identification and mapping of genetic loci affecting the free-threshing habit and spike compactness in wheat (Triticum aestivum L.). Theor. Appl. Genet. 2004, 108, 261–273.
- Elias, E.; Steiger, D.; Cantrell, R. Evaluation of lines derived from wild emmer chromosome substitutions: II. Agronomic traits. Crop Sci. 1996, 36, 228–233.
- Peng, J.; Ronin, Y.; Fahima, T.; Röder, M.S.; Li, Y.; Nevo, E.; Korol, A. Domestication quantitative trait loci in Triticum dicoccoides, the progenitor of wheat. Proc. Natl. Acad. Sci. USA 2003, 100, 2489–2494.
- Feldman, M.; Levy, A.A.; Fahima, T.; Korol, A. Genomic asymmetry in allopolyploid plants: Wheat as a model. J. Exp. Bot. 2012, 63, 5045–5059.
- Shaked, H.; Kashkush, K.; Ozkan, H.; Feldman, M.; Levy, A.A. Sequence elimination and cytosine methylation are rapid and reproducible responses of the genome to wide hybridization and allopolyploidy in wheat. Plant Cell 2001, 13, 1749–1759.
- Frankel, O.; Gerlach, W.; Peacock, W. The ribosomal RNA genes in synthetic tetraploids of wheat. Theor. Appl. Genet. 1987, 75, 138–143.
- Akhunova, A.R.; Matniyazov, R.T.; Liang, H.; Akhunov, E.D. Homoeolog-specific transcriptional bias in allopolyploid wheat. BMC Genom. 2010, 11, 505.
- Silva, M.; Pereira, H.S.; Bento, M.; Santos, A.P.; Shaw, P.; Delgado, M.; Neves, N.; Viegas, W. Interplay of ribosomal DNA loci in nucleolar dominance: Dominant NORs are up-regulated by chromatin dynamics in the wheat-rye system. PLoS ONE 2008, 3, e3824.
- Feldman, M.; Liu, B.; Segal, G.; Abbo, S.; Levy, A.A.; Vega, J.M. Rapid elimination of low-copy DNA sequences in polyploid wheat: A possible mechanism for differentiation of homoeologous chromosomes. Genetics 1997, 147, 1381–1387.
- Han, F.; Fedak, G.; Guo, W.; Liu, B. Rapid and repeatable elimination of a parental genome-specific DNA repeat (pGc1R-1a) in newly synthesized wheat allopolyploids. Genetics 2005, 170, 1239–1245.
- Kerber, E.; Green, G. Suppression of stem rust resistance in the hexaploid wheat cv. Canthatch by chromosome 7DL. Can. J. Bot. 1980, 58, 1347–1350.
- Akhunov, E.D.; Akhunova, A.R.; Anderson, O.D.; Anderson, J.A.; Blake, N.; Clegg, M.T.; Coleman-Derr, D.; Conley, E.J.; Crossman, C.C.; Deal, K.R. Nucleotide diversity maps reveal variation in diversity among wheat genomes and chromosomes. BMC Genom. 2010, 11, 702.
- Watanabe, N.; Sugiyama, K.; Yamagishi, Y.; Sakata, Y. Comparative telosomic mapping of homoeologous genes for brittle rachis in tetraploid and hexaploid wheats. Hereditas 2002, 137, 180–185.
- Simons, K.J.; Fellers, J.P.; Trick, H.N.; Zhang, Z.; Tai, Y.-S.; Gill, B.S.; Faris, J.D. Molecular characterization of the major wheat domestication gene Q. Genetics 2006, 172, 547–555.