Rapid diagnostic tests (RDTs), the unsung hero in malaria diagnosis, work to eliminate the prevalence of Plasmodium falciparum malaria through their efficient, cost-effective, and user-friendly qualities in detecting the antigen HRP2 (histidine-rich protein 2), among other proteins. However, the testing mechanism and management of malaria with RDTs presents a variety of limitations. SucTh ais entry discusses the parasitic factors that limit the performance of HRP2-based RDTs. By understanding the factors that affect the performance of HRP2-based RDTs in the field, researchers can work toward creating and implementing more effective and accurate HRP2-based diagnostic tools. Further research is required to understand the extent of these factors, as the rapidly changing interplay between parasite and host directly hinders the effectiveness of the tool.
- malaria
- RDTs
- HRP2
- Plasmodium falciparum
- diagnostics
1. Introduction
2. Parasitic Factors
2.1. HRP2 Persistence
Once HRP2 is released into the peripheral circulation during schizogony, it can persist for several weeks in the bloodstream, even when the infection is cleared, leading to false-positive RDT results [12][13][14][15][12,13,14,15]. Studies suggest that RDTs targeting HRP2 have low specificity for diagnosis of Pf malaria as significant yields of false-positive results have been observed, particularly in areas of high transmission [14][21][22][14,21,22]. Such results are attributed to the persistence of the protein in blood circulation even after the parasites have been cleared, which can lead to overmedication of patients and subsequent problems, particularly the emergence of drug-resistant or -tolerant parasite clones. The first study (1993) to describe HRP2 persistence found detectable antigen levels in the blood of 10 cases (25%) 5 days after treatment in a Tanzanian longitudinal cohort, 4 of which remained positive for 14 days [23]. Later studies detected circulating HRP2 for divergent time periods, ranging from a few days up to several weeks [14][22][24][25][14,22,24,25]. However, the kinetics of HRP2 antigenemia are not well understood, although important indicators, such as production rates and elimination half-life, have been estimated in both in vitro and in vivo studies [26][27][26,27]. A model of HRP2 kinetics was applied to clinical data from two studies on human infections, which indicated that in malaria naïve hosts, Pf parasites of the 3D7 strain produce 1.4 × 10−13 g of HRP2 per parasite for each replication cycle. In addition, it was demonstrated that the antigen’s persistence would cause the tests to stay positive for a minimum of seven days after treatment [28]. The duration of a positive RDT result after recommended treatment is mainly dependent on the density and duration of parasitemia before treatment, as well as the parasite-specific expression of HRP2. In one study, the mean duration of persistent antigenicity in Ugandan children who had a successful antimalarial treatment was 32 days [29]. This duration varied significantly depending on the pre-treatment parasitemia: patients with a parasite density >50,000/μL had, on average, persistent antigenicity for 37 days whereas patients with a density of <1000/μL had, on average, a persistent antigenicity of 26 days [29]. A more recent study looked further into the median half-life of HRP2 in blood by measuring HRP2 levels in individuals from Angola, Tanzania, and Senegal participating in therapeutic efficacy studies [30]. By fitting a first-order kinetics clearance model to the HRP2 concentration versus time (days), median clearance rate constants varied from 0.15 to 0.23 day−1 among countries, resulting in median half-lives of 4.5, 4.7, and 3 days in Angola, Tanzania, and Senegal, respectively. Such consistency in HRP2 clearance across different African regions suggests the presence of a common biological mechanism influencing HRP2 dynamics upon resolution of Pf infection [30]. It has also been hypothesized that the length of HRP2 positivity partly depends on the presence and affinity of host anti-HRP2 antibodies [28]. Highly sensitive RDTs (hsRDTs) claimed to have a 10-fold increase in sensitivity compared to the RDTs routinely available in the market [31]. Therefore, they provide the opportunity to more efficiently detect cases of malaria with low-parasitic densities and asymptomatic infections [32]. Traditional RDTs have a limit of detection (LOD) of approximately 800 pg/mL and show decreased sensitivity to detect low-density Pf infections (<100 parasites/µL). hsRDTs (with a LOD of 80 pg/mL) allow the detection of low-density Pf malaria infections (<100 parasites/µL), performing closely to expert microscopy (detection limit of 4–20 parasites/µL) [31]. However, this increased LOD can lead to a rise in the absolute number of false-positive results, as levels of persisting HRP2 that went undetected to date can now be picked up by hsRDTs [31][33][31,33]. In fact, a study utilizing participants from Angola, Tanzania, and Senegal countries demonstrated that the median duration of post-treatment RDT positivity increased from 13 to 16 days when utilizing the hsRDTs compared with conventional RDTs [30]. While the persistence of HRP2 can be seen as a drawback for malaria diagnosis and clinical management, it can also be used as a method to assess recent infections in non-infected people and consequently more accurately determine ongoing transmission [34][35][36][34,35,36]. Additionally, researchers have demonstrated that cases of recrudescence have different patterns in HRP2 concentrations over time. A study reported that infections which did not clear parasites due to treatment failure presented higher HRP2 levels at day 3 post-treatment compared with successfully treated infections, which suggests that these early changes in HRP2 levels could be used to monitor treatment success [30]. However, the field implementation of quantitative immunoassays, including the highly sensitive bead-based assay used in the above-mentioned studies [30][35][30,35], that allow antigen quantification appears logistically difficult, and therefore novel quantitative tools are needed if HRP2 is intended to be used as a marker for treatment prognosis.2.2. Variability of
P. falciparum
HRP2 and Homology with HRP3
The Pf genome encodes several proteins, including HRP2 and its structural homologue HRP3, that uncommonly contain repetitive sequences comprising a small handful of amino acids [9][37][38][9,37,38]. The mature configuration of HRP2 contains histidine-rich sequences that form the epitopes targeted by the mAbs in RDTs [9][18][39][9,18,39]. Past studies demonstrate that genetic variations (polymorphisms, gene deletions and duplications) have the potential to translate into different protein sequences [40]. HRP2 has been shown to be polymorphic in sequence composition of the repeated motif, leading to more than 30 sequence variants (or types) identified to date [38]. Additionally, there is significant polymorphism in both the number of repeated motifs and length between different parasite strains [9][38][9,38]. All considered, the question of whether such extensive diversity in HRP2 potentially affects the sensitivity of RDTs has been introduced, but few studies have since addressed it. The first study to observe extensive diversity in HRP2/3 sequences sequenced 75 P. falciparum lines and field isolates from 19 countries [9]. In this study, they identified a total of 18 unique amino acid repeats within the protein sequences of HRP2 and HRP3. Four of the repeat types (named 1, 2, 4 and 7) are shared by both proteins, repeat types 15–18 are specific for HRP3, and the remaining repeats are only present in HRP2. Of the 14 amino acid repeats identified in HRP2, only 5 were present in all isolates [9]. In addition, they showed variation in the frequency of repeats among different Pf isolates, as well as in the total number of repeats, its order within the sequence, and the number of each repeat within HRP2. These variations led them to identify 56 unique HRP2 sequences. Regarding the role such diversity could play on the performance of RDTs, they reported that isolates with higher numbers of type 2 and type 7 repeats were better recognized, particularly at low parasitemias (<250 parasites/µL). Remarkably, it was shown that the laboratory Pf D10 clone, lacking the pfhrp2 gene but positive for pfhrp3, could be detected by HRP2-based RDTs, suggesting that the candidate antigen for this cross-reactivity is HRP3. This aligns with a previous study that reported an 85–90% homology in nucleotide sequence-flanking repeats between pfhrp2 and pfhrp3 and an amino acid substitution of D/N in the major repeat of AHHAAD/N in a single parasite line [8], as well as with reported data on cross-reactivity between HRP2 and HRP3 for various HRP2-specific monoclonal antibodies [6]. In Madagascar Pf isolates, 93% (n = 13/14) of different HRP2 repeats identified in previous studies were detected in 229 successfully amplified pfhrp2 fragments, while for HRP3 protein, seven out of eight repeats were identified [41]. pfhrp2 sequence analysis predicted that 9% of Malagasy isolates would not be detected at parasite densities ≤250 parasites/μL [41]. The PfHRP2 and PfHRP3 antigens were found to be highly diverse in parasite isolates throughout Madagascar [41]. A later study identified new repeat sequences when 458 isolates from 38 countries were analyzed. These repeats (19–24) were specific to the HRP2 protein except for type 20, which was also present in the HRP3 protein [40]. Moreover, this study classified amino acid sequences for the HRP2 protein as Type A, B, or C depending on the product of the type 2 (AHHAHHAAD) and type 7 (AHHAAD) epitope repeats. Pf HRP2 Type A comprises the higher number of defined epitopes, the part of an antigen molecule to which an antibody attaches itself, followed by Type B and Type C. However, the predictability of RDT sensitivity using type 2 and type 7 repeats was not confirmed, as no correlation between detection rate using RDT and gene structure was identified, especially when the parasite density was <200 parasites/µL [40]. Studies in Pf isolates from India [39][42][43][44][39,42,43,44], the China-Myanmar border area [45], Myanmar [46], Yemen [37], Senegal [47][48][49][47,48,49], Mali [47], Uganda [47], French Guiana [50], Kenya [51][52][51,52], Mozambique [53], Tanzania [53], Nigeria [54], Sudan [38], Ethiopia [55], and Colombia [56] identified HRP2 and HRP3 sequences with different frequencies and number of repeats in each geographic area, although this trend was not identified in all cases. For example, repeat types 9, 11, 13, and 14 have rarely been observed in Africa [49]. However, all studies have described some similarities among isolates. One study [39] demonstrated that the amino acid sequences of HRP2 began with the type 1 repeat and ended with the type 12 repeat, and that repeat types 2 and 7 were found to be highly abundant. The same study also found a higher quantity of unique HRP2 protein sequences compared to HRP3 sequences. Furthermore, a study identified five novel repeats within HRP2 in Indian isolates [39]. In this same study, a relationship between the number of type 2 and type 7 repeats and RDT detection rate was statistically confirmed [39]. Similarly, a study in isolates from Senegal reported a mild association between the number of type 2 repeats and poor HRP2 RDT diagnosis (p = 0.046). The authors reported an increase in the number of isolates predicted to be non-sensitive to HRP2 RDTs based on HRP2 sequence for the years 2009 to 2011, suggesting the possibility of diagnostic selection pressure at play [49]. On the contrary, another study quantified HRP2 plasma levels by enzyme-linked immunosorbent assay (ELISA) in plasma samples with parasites of diverse pattern of repeat types and did not find a relationship between the number of repeats observed, or overall sequence length and HRP2 plasma concentrations [53]. Consistent with these findings, a study on Indian isolates observed no association between the number of type 2 and 7 repeats and the RDT performance to detect low-density infections [43]. In the same study, analysis of isolates led to the further identification of three and two new repeat sequences within HRP2 and HRP3, respectively, which resulted in a total of 34 repeat sequence variants. A study among Sudanese isolates also revealed that the RDT performance was not affected by a high-level of genetic diversity in HRP2 [38]. Recent studies in Kenyan [51][52][51,52] and Indian [44] isolates have also showed the presence of several new repeat sequences within HRP2 and HRP3. To summarize, the P. falciparum antigen HRP2 presents significant variation, but findings regarding the effect of such variability in RDT reactivity are divergent. This discordance among the several studies on the functional relevance of HRP2 repeats indicates that there is no established relationship between the RDT performance in the field and the number of repeats. However, it has been shown that recombinant HRP2 protein Type A is recognized with greater sensitivity in immunoassays compared with Types B and C [35]. Therefore, it is logical to think (but not easy to strongly prove) that some field isolates are more easily detected than others, particularly at low parasite densities.2.3.
P. falciparum hrp2
and
hrp3
Gene Deletions
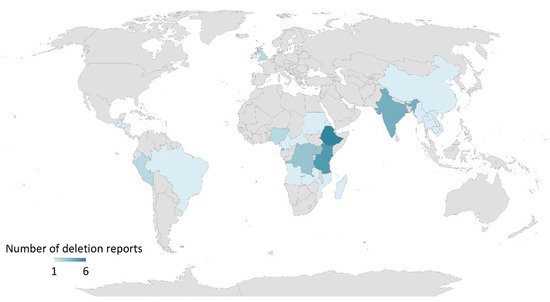
P. falciparum hrp2 and hrp3 genes do not only undergo extensive polymorphism but are also subject to deletions. Increasing false-negative HRP2-based RDT reports have rendered concerns, which have in turn led to the need for inclusion of other assays in malaria case management and elimination programs [10][11][10,11]. The first report on the presence of Pf isolates with pfhrp2 gene deletion came in 2010 from Peru [57], where eight among nine isolates collected in 2007 lacked the pfhrp2 gene and six lacked both pfhrp2 and pfhrp3 genes when tested by PCR. Plasma HRP2 levels were also not detectable by ELISA [57]. A retrospective analysis of 148 Pf isolates collected between 2003 and 2007 revealed that 41 and 70% of the isolates presented pfhrp2 and pfhrp3 gene deletions, respectively. In addition, 22% of the isolates lacked both pfhrp2 and pfhrp3 genes [57]. Another study from Peru also revealed that 20% of Pf isolates collected between 1998 and 2001 had a deleted pfhrp2 gene [58]. Increased pfhrp2 deleted Pf isolates (40%) were observed in samples collected from Peru between 2003 and 2005, suggesting that pfhrp2 deletion has occurred many times in the Pf isolates collected from this country [58]. The occurrence of pfhrp2 deletions is not just confined to South America but has also affected malaria endemic regions in Africa and Asia [10][11][10,11]. In 2010–2017, published reports of pfhrp2/3 deletions came from several countries in Africa (Mali, Mozambique, Senegal, Ghana, Kenya, Democratic Republic of Congo, Rwanda, Eritrea), Asia (India, China-Myanmar border), South America (Peru, Colombia, Brazil, French Guiana, Ecuador, Guyana, Suriname, Bolivia) and Central America (Honduras), which have been previously reviewed [10][11][10,11]. In 2018–2022, more reports from countries of African, Asian, South American and North American continents, as well as reports on travelers from the United Kingdom and Ireland, presented data on pfhrp2/3 deletions (Figure 1), Supplementary Table S1). These studies were identified in PubMed using the following search terms: (‘pfhrp2′, ‘pfhrp3′, and ‘deletion’) or (‘hrp2′, ‘hrp3′, and ‘deletion’) and publication dates from 01 January 2018 until the time of writing (14 July 2022). Based on these published reports, it can be concluded that Pf isolates with one or both pfhrp2/3 gene deletions are circulating throughout the world with varying prevalence in both high- and low-transmission areas. This scenario could adversely impact the life of an affected individual due to health consequences from delayed treatment or no treatment, while also affecting overall healthcare efforts in malaria case management and preventing ongoing transmission.