1. Introduction
The integration of metabolism with virulence has become a new paradigm in the host–fungal pathogen interaction. In addition to virulence attributes, the term “fitness attributes” has been defined as the cellular functions that are required to support microbial growth and survival
[1]. The fitness attributes include the metabolic capacity to assimilate host nutrients, resistance to host-imposed stress, tolerance to elevated host temperature, and the construction of a robust cell wall
[2]. The inactivation of fitness attributes will diminish the ability of a fungus to obtain nutrients or combat environmental stressors and hence, will attenuate its ability to grow, express virulence factors, and ultimately cause infection. The metabolism provides a platform for generating the precursors and energy required for growth, antioxidant production and cell wall remodeling. Moreover, metabolic adaptation can modulate the expression of virulence factors and immunogenicity. Thus, the metabolism impacts fungal pathogenicity through both virulence and fitness attributes and is indispensable for a fungal pathogen to colonize and infect a host successfully.
Fungi can infect multiple sites on the human body and cause both superficial and life-threatening infections. As one and a half million people are killed by pathogenic fungi every year, fungal pathogens are known as a “hidden killer”
[3]. The most common culprits of human fungal infection come from four major fungal groups:
Candida species,
Cryptococcus neoformans,
Aspergillus fumigatus and thermal dimorphic fungi
[4]. Important human fungal pathogens belonging to the thermal dimorphic fungus group are
Talaromyces marneffei,
Histoplasma capsulatum,
Coccidioides immitis,
Paracoccidioides brasiliensis,
Blastomyces dermatitidis and
Sporothrix schenckii. During infection, the host’s innate immune cells, such as macrophages or neutrophils, commonly phagocytize and destroy the fungal cells by generating reactive oxygen species (ROS) and reactive nitrogen species (RNS)
[5]. The host macrophages have been reported to produce up to 14 mM hydrogen peroxide and up to 57 uM nitric oxide in response to fungal infections
[6][7][6,7]. To cope with these host-imposed stressors, fungal pathogens possess antioxidant defense systems with both enzymatic and non-enzymatic mechanisms
[8]. In addition, several fungi can switch their morphology to protect themselves from the human immune system.
Morphological plasticity is one of the main virulence attributes in pathogenic fungi. Cell differentiation and development contribute to diverse morphological changes, including germination, conidiation, morphological switching between yeast and mold forms, and even autolysis. Fungal species generally undergo a morphological transformation during host colonization by responding to specific environmental cues. For example, thermal dimorphic fungi switch morphology from a multicellular mold in environmental niches to a yeast form in warm-blooded hosts due to temperature changes
[9][10][11][9,10,11]. Another example is
Candida albicans, which can be a commensal organism of the human microbiome while also being the most prevalent human fungal pathogen. At least nine distinct cell shapes have already been found in this species.
C. albicans changes morphotypes when it inhabits different host niches or when it changes between being a commensal or pathogenic organism
[12]. In
A.
fumigatus, a ubiquitous pathogenic mold, germination of conidia and hyphal growth occurs during an invasive infection of human lungs, while conidiation is strictly inhibited
[13][14][15][13,14,15]. The dysregulation of these morphology pathways consistently attenuates the virulence of the pathogenic fungi in animal studies
[15][16][17][18][15,16,17,18]. Overall, cell differentiation and development are critical for fungal morphogenesis and pathogenicity.
2. Glutathione Systems
Glutathione (L-γ-glutamylcysteinylglycine) is a crucial metabolite in eukaryotes and plays a major role in protecting cells against oxidative damage
[19][20][19,20]. Glutathione directly scavenges diverse oxidants, such as superoxide anion, hydroxyl radical, nitric oxide and carbon radicals and is also a cofactor for various antioxidant enzymes, including glutathione peroxidases and glutathione S-transferase
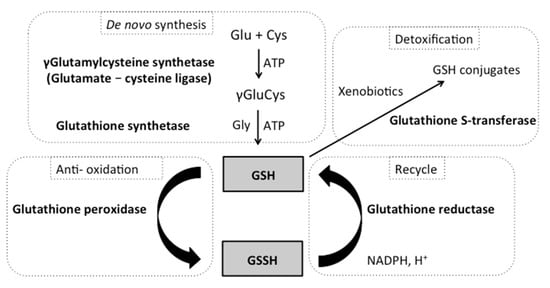
[21][22][21,22]. There are two states of glutathione in the cells: reduced glutathione (GSH) and oxidized glutathione disulfide (GSSG) (
Figure 1). Importantly, GSH is a major tissue antioxidant, while GSSG is accumulated when cells are exposed to increased levels of oxidative stress. Thus, increased ratios of GSSG to GSH are indicative of oxidative stress, and cells tightly maintain levels of reduced glutathione through the balance of its synthesis and reduction.
Figure 1. The glutathione pathway in Saccharomyces cerevisiae. The synthesis of glutathione (GSH) is a two-step process catalyzed by the enzyme γ-glutamylcysteine synthetase (GSH1) and glutathione synthetase (GSH2) in the cytoplasm. GSH plays a crucial role in the protection of macromolecules from stressors, especially oxidants. The reduced form (GSH) directly scavenges diverse oxidants and xenobiotics via the action of glutathione peroxidase (GPX1-3) and glutathione S-transferase (GST) and becomes oxidized (GSSH). Regeneration of oxidized GSSG to reduced GSH is catalyzed by glutathione reductase (GLR1) (requires NADPH).
The GSH/GSSH pathway is composed of five enzymes, which will be the major focus of this review paper. The first two enzymes are involved with glutathione de novo biosynthesis via two ATP-dependent steps: the first step is catalyzed by γ-glutamylcysteine synthetase (GSH1), while the second step is catalyzed by glutathione synthetase (GSH2) (Figure 1). Next, glutathione peroxidase and glutathione S-transferase catalyze the production of GSSH. Glutathione peroxidase is a major enzyme in the defense against hydrogen peroxide (GPx: 2GSH + ROOH → GSSG + H2O + ROH, EC 1.11.1.9). Glutathione S-transferase is involved with the detoxification of many xenobiotic compounds by catalyzing the conjugation of substrates to GSH (GST: GSH + RX → GS-X + RH, EC 2.5.1.18), which can then be eliminated from the cells via glutathione conjugate pumps. Lastly, glutathione reductase catalyzes the regeneration of GSH (GSSG + NADPH + H+ → 2GSH + NADP+, EC 1.6.4.2).
3. Glutathione and Role in Oxidative Stress Protection
Molecular mechanisms elucidating how the glutathione system plays a crucial role in the fungal stress response are primarily obtained from model fungi, such as Saccharomyces cerevisiae, Schizosaccharomyces pombe and Aspergillus nidulans, or common human fungal pathogens, such as C. albicans, A. fumigatus and C. neoformans. In thermal dimorphic fungi, however, an understanding of the glutathione system is lacking because glutathione genetic studies have not been extensively explored, as seen in model fungi and other human fungal pathogens; the role of the glutathione system is mostly inferred from gene or protein expression analyses. Therefore, reswearchers summarized the experimentally verified functions of each homologous glutathione gene from diverse fungal species (Table 1). This summarized table will be beneficial in predicting the gene functions for other less studied fungi, such as thermal dimorphic fungi or other non-model fungi.
3.1. γ-Glutamylcysteine Synthetase and Glutathione Synthetase
As sTh
own in Table 1, the deletion of the genes encoding glutathione synthetic enzymes (
GSH1 or
GSH2 genes or their homologs) resulted in fungal mutants that were glutathione auxotrophs and had an increased sensitivity to various oxidants. Interestingly, the deletion of
gshA in
A. fumigatus also impaired cellular iron sensing, indicating crosstalk between glutathione biosynthesis and fungal iron homeostasis
[23]. Likewise, the deletion of glutathione synthetase (
GSH2) from
C. neoformans resulted in glutathione auxotrophy under iron starvation-induced stress
[24]. Indeed, glutathione is proposed to be involved with iron metabolism by its requirement in the Fe–S cluster assembly
[25][26][25,26]. Importantly, the
GSH1 and
GSH2 genes are essential in diverse fungal species. A study in
H. capsulatum reported that the deletion of the
GSH1 or
GSH2 genes caused non-viable mutants, indicating that both the
GSH1 and
GSH2 genes are essential. Consistent with the result from
H. capsulatum,
gshA (γ-glutamylcysteine synthetase gene) from
Aspergillus oryzae was also reported to be an essential gene, while the role of the glutathione synthetase gene (
GSH2 homolog) in cellular viability has not been characterized in this fungal species
[27]. In
Candida glabrata,
GSH1, but not
GSH2, was an essential gene. These results suggest that the role of the
GSH1 and
GSH2 genes differs from various species in cell viability, and glutathione synthesis has an essential role in iron homeostasis in many fungi.
Table 1. Analyses of glutathione-related enzymes in diverse fungal species.
|
Enzymes in glutathione system
|
Species
|
Gene name
|
Phenotypes
|
References
|
|
γ-glutamylcysteine synthetase
|
Saccharomyces cerevisiae
|
GSH1
|
The gsh1Δ mutant showed glutathione auxotrophy, slower growth and increased sensitivity to oxidative stress.
|
[28,29]
|
|
Schizosaccharomyces pombe
|
gcs1
|
- The gcs1Δ mutant showed glutathione auxotrophy and sensitivity to cadmium.
- The gcs1Δ mutant was unable to sporulate.
|
[30,31]
|
|
Candida albicans
|
GCS1
|
- The gcs1Δ mutant showed glutathione auxotrophy, increased ROS production and apoptosis.
- The gcs1Δ mutant showed no change in morphogenesis and virulence.
|
[32,33]
|
|
Nakaseomyces glabrataa (formerly, Candida glabrata)
|
GSH1
|
- The gsh1Δ mutant was lethal.
- A conditional deletion mutant, gsh1Δpro2-4, showed low glutathione levels and slower growth in media lacking glutathione.
- The gsh1Δpro2-4 mutant showed sensitivity to oxidative stress (H2O2, menadione) and cadmium.
|
[33,34]
|
|
Histoplasma capsulatum
|
GSH1
|
- The GSH1 gene was expressed only in the yeast form.
- The gsh1Δ mutant was lethal.
- The GSH1 overexpression mutant showed an inability to switch from yeast to mold form.
|
[35]
|
|
Glutathione synthetase
|
Saccharomyces cerevisiae
|
GSH2
|
The gsh2Δ and the GSH2 overexpression mutants showed normal responses to oxidative stress.
|
[36]
|
|
Schizosaccharomyces pombe
|
gsh2
|
The gsh2Δ mutant showed glutathione auxotrophy and sensitivity to cadmium.
|
[30,31]
|
|
Nakaseomyces glabrataa (formerly, Candida glabrata)
|
GSH2
|
- The gsh2Δ mutant showed glutathione auxotrophy.
- The gsh2Δ mutant showed low glutathione levels and sensitivity to oxidative stress (H2O2, menadione) and cadmium.
- The gsh2Δ mutant showed resistance to tert-butyl hydroperoxide and cumene hydroperoxide stressors.
|
[34]
|
|
Cryptococcus neoformans
|
GSH2
|
- The gsh2Δ mutant showed glutathione auxotrophy under iron starvation conditions.
- The gsh2Δ showed low glutathione levels and sensitivity to the oxidative stressor diamide, but not H2O2.
- The gsh2Δ mutant showed sensitivity to a high salt stressor, the cell wall damaging agent Congo red, and antifungal drugs.
- The gsh2Δ showed impairment in virulence-related traits, including defects in capsule formation, melanin production and growth at 37 °C.
|
[24]
|
|
Histoplasma capsulatum
|
GSH2
|
- The GSH2 gene was highly expressed in the yeast form.
- The GSH2 overexpression mutant showed an inability to switch from yeast to mold form.
- The gsh2Δ mutant was lethal.
|
[35]
|
|
Glutathione reductase
|
Saccharomyces cerevisiae
|
GLR1
|
The glr1Δ mutant showed sensitivity to oxidative stress (H2O2).
|
[37,38]
|
|
Schizosaccharomyces pombe
|
pgr1
|
- The pgr1 overexpression mutant showed resistance to the oxidative stressor menadione but not H2O2.
- The pgr1 gene expression was induced by various oxidative stressors (menadione, cumeme hydroperoxide and diamide, but not H2O2), high salt levels (NaCl), high temperatures and starvation.
- The pgr1Δ mutant was lethal.
|
[39]
|
|
Candida albicans
|
GLR1
|
- The glr1Δ mutant showed sensitivity to oxidative stress (H2O2) but not formaldehyde or nitrosative stress (NO).
- The glr1Δ mutant showed an inability to detoxify GSSG.
- The glr1Δ mutant showed impairment in macrophage killing.
- The glr1Δ mutant showed decreased virulence, while the GLR1 overexpression mutant showed increased virulence.
|
[40]
|
|
Cryptococcus neoformans
|
GLR1
|
- The glr1 gene expression was induced by nitric oxide (NO).
- The glr1Δ mutant showed normal morphology.
- The glr1Δ mutant showed sensitivity to nitric oxide stress but not peroxide stress.
- The glr1Δ mutant became avirulent in an inhalation model of mouse infection and showed sensitivity to macrophage killing.
|
[41]
|
|
Aspergillus nidulans
|
glrA
|
- The glrAΔ mutant showed slower growth under normal conditions.
- The glrAΔ mutant showed defects in conidia germination at high temperatures.
- The glrAΔ mutant showed sensitivity to various oxidants (menadione, diamide and H2O2).
- The glrAΔ mutant accumulated a less reduced form of GSH, more intracellular ROS, and had decreased respiration activity.
|
[42]
|
|
Paracoccidioides brasiliensis
|
GR
|
The vPb18 virulent strain showed increases in both levels of the GR gene and enzymatic activity.
|
[43]
|
|
Glutathione peroxidase
|
Saccharomyces cerevisiae
|
GPX1-3
|
- GPX1-3 genes encoded for phospholipid hydroperoxide glutathione peroxidase.
- The GPX3 product was a major glutathione peroxidase.
- The GPX3 gene was constitutively expressed.
- The GPX1 gene expression was induced under glucose starvation.
- The GPX2 gene expression was induced by many oxidative stressors.
- The gpx3Δ mutant showed sensitivity to peroxides (H2O2 and tert-butyl hydroperoxide).
- The gpx1Δ and gpx2Δ mutants showed no sensitivity to oxidative stress.
- The gpx1Δgpx2Δgpx3Δ mutant showed sensitivity to H2O2 and phospholipid hydroperoxide (polysaturated fatty acid linolenate 18:3).
|
[44,45]
|
|
Candida albicans
|
GPX3
(ScGPX1 homolog)
|
- The gpx3Δ mutant (orf19.4436Δ) showed sensitivity to H2O2 and was defective in hyphal formation within macrophage cells.
- The gpx3Δ mutant showed impairment in killing macrophages and Galleria mellonella.
- The gpx3Δ mutant showed normal virulence in a murine model of infection.
|
[46,47]
|
|
GPX31-
33
(ScGPX3/
HYR1 homolog
|
- The GPX31 is a major glutathione peroxidase.
- The gpx31Δ (orf19.86Δ) and the gpx31Δgpx32Δgpx33Δ mutant (orf19.86Δorf19.85Δorf19.87Δ) showed sensitivity to oxidative stressors (H2O2 and t-butylhydroperoxide but not menadione), UV light, heavy metals (cadmium and silver), and cell wall stressors (congo red and calcofluor white).
|
|
Cryptococcus neoformans
|
GPX1, GPX2
|
- GPX1 and GPX2 gene expressions were induced in response to t-butylhydroperoxide and cumene hydroperoxide and repressed in response to nitric oxide.
- GPX2 gene expression was induced in response to the hydrogen peroxide stressor.
- The gpx1Δ and gpx2Δ mutants showed normal morphology, melanin production and capsule formation.
- The gpx1Δ and gpx2Δ mutants showed sensitivity to cumene (hydroperoxide) but not superoxide, hydrogen peroxide or nitric oxide stressors.
- The gpx2Δ mutant showed higher sensitivity to cumene hydroperoxide than the gpx1Δ mutant at high concentrations.
- The gpx1Δ mutant, but not the gpx2Δ mutant, showed sensitivity to the peroxide stressor t-butylhydroperoxide.
- The gpx1Δ and gpx2Δ mutants showed sensitivity to macrophage killing, yet the mutants were still virulent in a mouse model.
|
[48]
|
|
Aspergillus fumigatus
|
hyr1 (ScGPX3/
HYR1 homolog)
|
- hyr1 gene expression was upregulated in hyphae and conidia when exposed to neutrophils.
- The hyr1 gene expression was induced when exposed to H2O2.
|
[49,50]
|
|
Talaromyces marneffei
|
gpx1 (ScGPX3/HYR1 homolog)
|
- Gpx1 is an antigenic protein.
- gpx1 gene expression was upregulated in the yeast form.
|
[51,52]
|
|
Glutathione S- transferase
|
Saccharomyces cerevisiae
|
GTT1-2
|
- GTT1 gene expression was induced during the diauxic shift and stationary phase.
- The gtt1Δ, gtt2Δ, and gtt1Δgtt2Δ showed sensitivity to heat shock in a stationary phase and slower growth at a high temperature of 39 °C.
- The grx1Δgrx2Δgtt1Δgtt2Δ mutant showed sensitivity to xenobiotics (1-chloro-2,4-dinitrobenzene), heat and the oxidative stressors (cumene hydroperoxide and H2O2).
|
[53,54]
|
| |
Schizosaccharomyces pombe
|
gst1-3,
|
- The gst1Δgst2Δ and gst3Δ mutants showed sensitivity to peroxide stressors (H2O2 and t-butylhydroperoxide) and the antifungal drug fluconazole.
- The gst1Δgst2Δ and gst3Δ mutants showed resistance to the peroxide stressor diamide.
- gst1, gst2, and gst3 gene expressions were induced during the stationary phase and in response to hydrogen peroxide.
- All Gst1, 2 and 3 enzymes have glutathione transferase activity, and the Gst3 enzyme also has glutathione peroxidase activity.
|
[55]
|
| |
Candida albicans
|
GST2
|
- The gst2Δ mutant showed sensitivity to oxidative stress (H2O2).
- GST2 gene expression was induced under nitrogen limitation.
- The gst2Δ mutant showed defects in hyphal switching under nitrogen starvation-induced filamentous growth.
|
[56]
|
| |
Aspergillus nidulans
|
gstA
|
- The gstAΔ mutant showed sensitivity to the oxidant diamide, the fungicide carboxin, various xenobiotics (pyrrolnitrin and sulphanilamide), and heavy metals (selenium, silver and nickel).
- The gstAΔ mutant showed normal growth in the presence of 1-chloro-2,4-dinitrobenzene.
- The gstA gene expression was induced in response to xenobiotics (1-chloro-2,4-dinitrobenzene) and oxidative stress (H2O2).
|
[57]
|
| |
Aspergillus fumigatus
|
gstA, gstB, gstC
|
- All gstA, B and C enzymes have both glutathione transferase and glutathione peroxidase activities.
- The gstA and gstC genes were constitutively expressed under normal conditions, and their expression levels were inducible in response to oxidative stress (H2O2).
- The expression of all gst genes was induced in response to xenobiotics (1-chloro-2,4-dinitrobenzene).
|
[58]
|
| |
Paracoccidioides brasiliensis
|
GST1-3
|
The vPb18 virulent strain showed increased levels of the GST1-3 genes.
|
[43]
|
| |
Paracoccidioides lutzii
|
GST
|
GST was exclusively secreted in the yeast form.
|
[59]
|
3.2. Glutathione Reductase
In addition, glutathione reductase is required for resistance to oxidative stress because the deletion of the glutathione reductase gene commonly results in fungal mutants that are sensitive to various stressors. The details of the growth and stress response defects in deletion mutants are different according to the species. For example, in
S. pombe yeast, glutathione reductase is indispensable for growth, as the
pgr1Δ strain was not viable due to the accumulation of GSSG
[28][29][39,60]. In
S. cerevisiae and
C. albicans, the
grl1Δ mutant was viable and only showed growth defects under oxidative stress
[30][31][32][37,38,40]. In
C. neoformans, the
gr1Δ mutant grew normally under normal conditions and was sensitive to only nitric oxide stress but not to peroxide stress
[33][41]. In
A. nidulans, the
grlAΔ mutant exhibited growth defects, even under normal conditions, yet was still viable
[34][35][42,61]. The
grlAΔ strain of
A. nidulans was also defective in its growth under high temperatures. Overall, these results suggest that glutathione reductase functions differently among fungal species.
3.3. Glutathione Peroxidase
Furthermore, glutathione peroxidase plays a crucial role in protecting fungi against oxidative stress since the absence of glutathione peroxidase gene(s) results in fungal strains that are not able to cope with various oxidants, especially peroxides. Nonetheless, distinct cellular responses to oxidative stress could be observed among fungal species. For example, the
gpx3Δ mutants from
S. cerevisiae and
C. albicans were highly sensitive to H
2O
2, while the
gpx1Δ and
gpx2Δ mutants from
C. neoformans were not sensitive to H
2O
2 but were sensitive to other peroxides. Furthermore, the
gpx1Δ and
gpx2Δ mutants from
S. cerevisiae and
C. albicans did not show any defects in response to oxidative stress, and hence,
GPX3 is proposed to be the main gene encoding for glutathione peroxidase in these fungi. In
T. marneffei, the glutathione peroxidase gene (
gpx1; the homolog of the glutathione peroxidase
HYR1 gene from
S. cerevisiae) was isolated as one of the antigenic proteins
[36][51]. The expression levels of the
T. marneffei gpx1 gene were high in the pathogenic yeast form and were relatively unchanged in the conidia or mold forms. These results imply that glutathione peroxidase contributes to immunological response during
T. marneffei infection and plays an important role in the pathogenic yeast phase. Collectively, these results suggest that glutathione peroxidase is required for the general oxidative stress defense mechanisms yet could respond distinctly to the different stressors, depending on the fungal species.
3.4. Glutathione S-Transferase
Glutathione S-transferase is involved in the resistance to xenobiotics because this enzyme can detoxify a broad range of harmful substances. In addition, glutathione S-transferase can also protect the cells against oxidative stress as it possesses GSH-dependent peroxidase activity. Accordingly, the glutathione S-transferase gene deletion mutants from a wide range of fungi became sensitive to both xenobiotics and various stressors. As seen in the case of other glutathione-related genes, there are differences in the glutathione S-transferase function among individual fungal species. In
S. pombe, the
gst1Δ
gst2Δ and
gst3Δ mutants were sensitive to the antifungal drug fluconazole, suggesting the role of glutathione S-transferase in mediating drug resistance. In
C. albicans, the
GST2 gene was additionally induced under nitrogen starvation
[37][56]. In
S. cerevisiae, the
gtt1Δ,
gtt2Δ, and
gtt1Δ
gtt2Δ mutants showed an increased sensitivity to heat shock or exhibited growth defects at high temperatures. In
A. nidulans, the
gstAΔ mutant was sensitive to heavy metals
[38][57]. Taken together, glutathione S-transferase is an important enzyme that protects fungal cells from diverse types of substances and stressors.