2. Cyclic Lipopeptides
Cyclic lipopeptides (CLPs) are common secondary metabolites isolated from marine-derived
Bacillus. The CLPs are a class of metabolites with structural diversity produced by multifarious bacterial genera
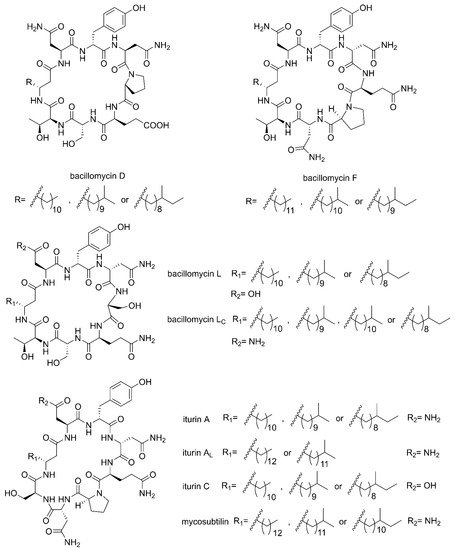
[39][42]. There are three families of CLPs being of particular importance, namely surfactins, iturins and plipastatins, all consisting of a short cyclic oligopeptide linked to the tail of a fatty acid
[14]. Surfactin sequences comprise of seven amino acids and a
β-hydroxy fatty acid chain containing 12–16 carbons
[40][43]. The iturin family sequences are composed of heptapeptides and a
β-amino fatty acid chain of 14–17 carbon atoms, which consists of bacillomycin D, F, L, Lc, iturin A, A
L, C and mycosubtilin (
Figure 1)
[41][44]. The plipastatin family comprise of ten amino acids and a
β-hydroxy fatty acid containing 14–18 carbon atoms
[42][45].
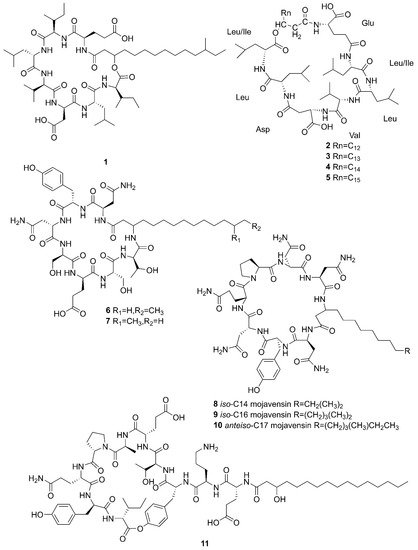
Figure 2 lists the structures of cyclic lipopeptides produced by marine-derived
Bacillus species.
Figure 1. The structures of iturin family members (bacillomycin D, F, L, Lc, iturin A, AL, C and mycosubtilin).
Figure 2. Cyclic lipopeptides produced by marine-derived Bacillus species.
Compounds
1–5 belong to surfactin family. A new CLP surfactin named
anteiso-C
15 Ile
2,7 surfactin (
1) was isolated from
B.
velezensis SH-B74 in the China Center for Type Culture Collection (CCTCC), which collected from the marine sediments, comprising of an
anteiso-C
15 type saturated fatty acid chain, and a peptidic backbone of L-Glu
1, L-Ile
2, D-Leu
3, L-Val
4, L-Asp
5, D-Leu
6, L-Ile
7 [43][46]. Rn-Glu
1-Leu/Ile
2-Leu
3-Val
4-Asp
5-Leu
6-Leu/Ile
7 (
2–5) belonging to surfactin homolog were isolated from
B.
licheniformis MB01 collected from sediments in the Bohai Sea, China
[44][47]. Compounds
6–10 belong to the iturin family. A novel lipopeptide antibiotic bacillopeptin named bacillopeptin B
1 (
6) and a known compound, bacillopeptin B (
7) were detected in the fermentation broth of a marine sediment-derived
B. amyloliquefaciens SH-B74 collected from sediments in the South China Sea. More precisely, compound
6 as a member of bacillopeptin family has the same amino-acid sequence and the same molecular weight as compound
7, but has a different fatty-acid residue
[22]. Compounds
8–10 were characterized as cyclic lipopeptides with saturated
β-amino fatty acid chain residues,
iso-C14 mojavensin,
iso-C16 mojavensin, and
anteiso-C17 mojavensin, all of which were produced by a marine-derived
B. mojavensis B0621A obtained from the mantle of a pearl oyster
Pinctada martensii in the South China Sea
[45][48]. In addition, plipastatin A1 (
11), belonging to the plipastatin family, was obtained by solidphase extraction and reversed-phase high-performance liquid chromatograph (RP-HPLC) from the fermentation broth of a marine sediment-derived
B. amyloliquefaciens SH-B74 in the CCTCC
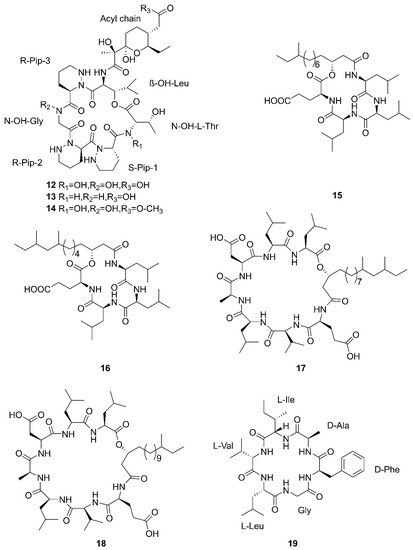
[46][49]. A new cyclic hexapeptide with three piperazic acids (N-OH-Thr, N-OH-Gly,
β-OH-Leu) named dentigerumycin E (
12) and two reported derivatives, 2-N, 16-N-deoxydenteigerumycin E (
13) and dentigerumycin E methyl ester (
14), were isolated from coculture of marine
Streptomyces and
Bacillus strains collected together from the intertidal mudflat in Wando, Republic of Korea. It is worth mentioning that only compound
12 showed antiproliferative and antimetastatic activities against human cancer cells, suggesting that 2-N-OH, 16-N-OH, and 37-OH (carboxylic acid) are essential for the activities
[25]. Two novel cyclic lipopeptides, bacilotetrin A (
15) and bacilotetrin B (
16), possessing three leucines and one glutamic residue cyclized with a lipophilic 3-hydroxyl fatty acid, were isolated from
B. subtilis 109GGC020 in the sediments from the Gageocho of southern reef, Republic of Korea
[12]. Additionally, gageopeptins A (
17) and B (
18), two novel cyclic lipopeptides, were isolated from the same strain in the same sediments as above
[47][50]. A new cyclic hexapeptide named bacicyclin (
19) was purified from
Bacillus sp. BC028 associated with the blue mussel
Mytilus edulis collected from the western shore of the Baltic Sea in Germany
[48][51].
3. Diketopiperazines
Cyclicpeptide diketopiperazines consist of residues of two amino acids and mevalonic acid
[49][52].
Figure 3 lists the structures of diketopiperazines that were produced by marine-derived
Bacillus species.
Figure 3. Diketopiperazines produced by marine-derived Bacillus species.
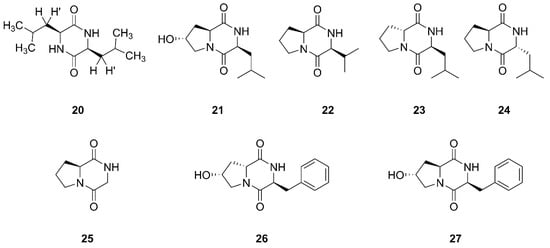
Compound
20 was established as a diketopiperazine (3S, 6S)-3,6-diisobutylpiperazine-2,5-dione, which was isolated from the ethyl acetate extract of the culture broth of
Bacillus sp. SPB7. This strain SPB7 was obtained from marine sponge
Spongia officinalis collected from the Palk Bay of Bengal, India. In particular, this is the first time that compound
20 had been isolated from a sponge-associated microbe
[50][53]. Finally, seven cyclic dipeptides compounds
21–
27 were identified and characterized as cyclo (L-leu-trans-8-hydroxy-L-pro), cyclo (L-val-L-pro), cyclo (D-pro-L-leu), cyclo (L-pro-D-leu), cyclo (gly-L-pro), cyclo (L-phe-cis-8-hydroxy-D-pro), and cyclo (L-phe-trans-8-hydroxy-L-pro), respectively, which were isolated from
Bacillus sp. UST050418-715 collected from sponge in the sea near St. Juan Island, Washington, USA
[51][54]. Compound
25 was also found from sponge-endosymbiotic
Bacillus species collected from Agatti island located in the Arabian Sea, in the Laksha Archipelago of India
[52][55].
4. Linear Lipopeptides
Linear lipopeptide is a kind of lipopeptide, in which amino acids are connected in turn into linear, unconnected head and tail and no cyclic structure. Fatty acids are connected to α-amino groups or other hydroxyl groups at the N-terminal of the peptide chain
[53][56].
Figure 4 lists the structures of linear lipopeptides that were produced by marine-derived
Bacillus species.
Figure 4. Linear lipopeptides produced by marine-derived Bacillus species.
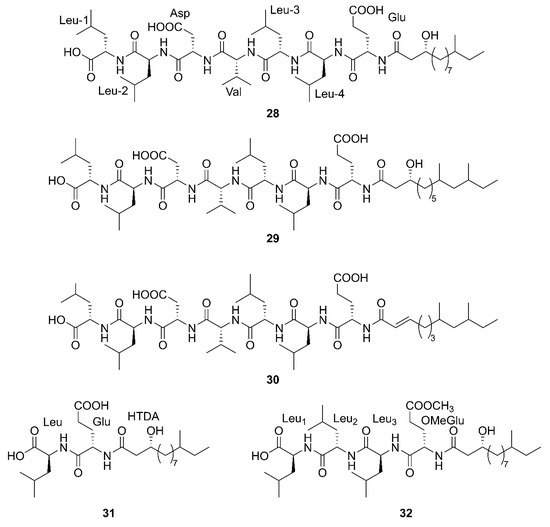
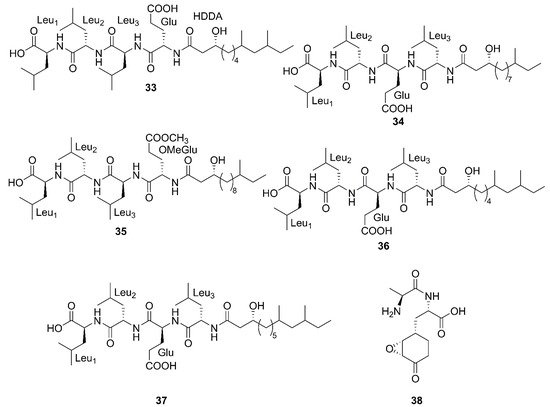
Three newfound linear lipopeptides named gageostatins A (
28), B (
29) and C (
30), comprising of hepta-peptides and new 3-
β-hydroxy fatty acids yielded by a marine-derived bacterium
B. subtilis from the culture broth
[35]. Furthermore, three novel linear lipopeptides possessing di- and tetrapeptides and a new fatty acid, gageotetrins A (
31), B (
32) and C (
33), were isolated from a marine
B. subtilis [54][57]. Four unreported lipopeptides, gageopeptides A (
34), B (
35), C (
36), and D (
37) were isolated and identified from a marine-derived bacterium
B. subtilis, which consisted of tetrapeptides and 3-
β-hydroxy fatty acids
[55][58]. The fatty acid of
28,
31,
32 and
34 was identical and determined as a 3-
β-hydroxy-11-methyltridecanoic acid. Likewise, compounds
29 and
37 both possessed the same fatty acid, 3-
β-hydroxy-9,11-dimethyltridecanoic acid. Moreover, the fatty acid unit of
33 and
36 was 3-
β-hydroxy-8,10-dimethyldodecanoic acid. In particular, the absolute stereochemistry at C-3 of the fatty acids of linear lipopeptides
28–
37 is
R configuration except
30. Additionally, the configuration of the amino acid residues in
28–
37 was found to be
L-form, while Val in
28–
30 was
D-form. Besides, bacilysin (
38), another identified dipeptide, was isolated from seaweed-associated
B. amyloliquefaciens MTCC 10456 in Microbial Type Culture Collection and Gene Bank (MTCC) of Chandigarh in India. Notably, this is the first report on the co-production and isolation of anti-
Malassezia spp. chemicals from marine
Bacillus species
[41][44]. In conclusion, all linear Lipopeptide mentioned above were obtained from the Gageocho in the southern reef (Republic of Korea) except
38.
5. Nonribosomal Peptides
Nonribosomal peptides (NRPs) are large enzyme complexes with a modular structure responsible for binding a particular amino acid. NRPSs of
Bacillus are synthesized by large multimodular nonribosomal peptide-synthetase (NRPS) through prolonging the active monomers of amino acid building blocks
[56][59].
Figure 5 lists the structures of nonribosomal peptides produced by marine-derived
Bacillus species.
Figure 5. Nonribosomal peptides produced by marine-derived Bacillus species.
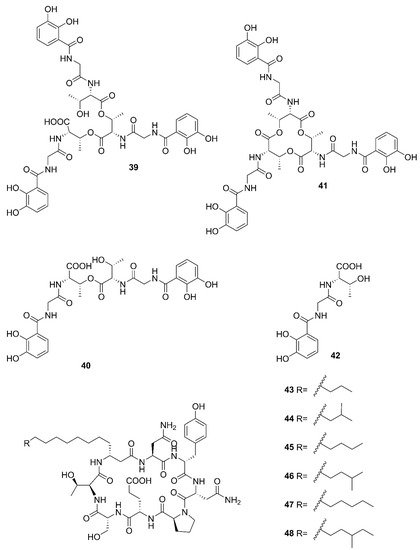
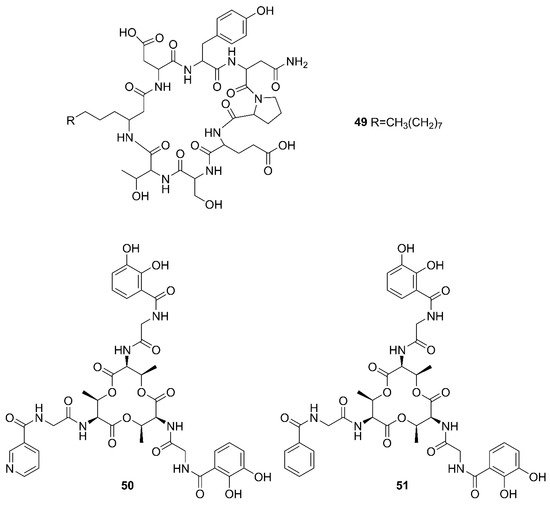
Two unreported compounds, bacillibactin B (
39) and bacillibactin C (
40), along with the known compounds Bacillibactin (
41) and S
VK21 (
42), were discovered from
Bacillus sp. named PKU-MA00093 from sponges, corals and sediments in the South China Sea. Additionally, compounds
43–
48 were characterized as bacillomycin D,
iso-C15 bacillomycin D, C15 bacillomycin D,
iso-C16 bacillomycin D, C16 bacillomycin D, and
anteiso-C17 bacillomycin D, respectively. They were isolated from
Bacillus sp. PKU-MA00092 collected from sponges, corals and sediments in the South China Sea. Notably, this was the first time to report the structures of
45 and
47 with fully specified
1H NMR and
13C NMR data; their structures are highly similar except for the fatty acid moieties
[57][60]. Compounds
45 and
47 in company with C14 bacillomycin D (
49), were obtained from seaweed-associated
B. amyloliquefaciens MTCC 10456 collected from seaweed in the MTCC, of Chandigarh, India
[41][44]. Moreover, compounds
45 and
49 were also isolated from the methanol extract harvested from marine-derived
B. megaterium CGMCC7086 obtained from the intestines of marine fish in the Yellow Sea of East China by two-step ultrafiltration and liquid chromatography-electronic spray ionization-tandem mass spectrometry (LC-ESI-MS/MS). Using a highly-efficient separation technique and identification method, more than 40 lipopeptides variants were identified from a
Bacillus strain
[23]. Besides, compound
47 was isolated from
B. subtilis B38 strain
[58][61]. Two unique bacillibactins, bacillibactins E (
50) and F (
51) were the first bacterial siderophores containing nicotinic and benzoic acid moieties isolated from a marine sponge
Cinachyrella apion associated
Bacillus sp. WMMC1349 collected from the west shore of Ramrod Key in Florida
[24].
6. Polyketides
Polyketides are a class of extremely large secondary metabolites assembled from simple acyl-coA compounds
[59][62].
Bacillus species of marine origin was a potential source of bioactive compounds of polyketides and bacteriocins with significant antimicrobial activity against human pathogens
[60][63].
Figure 6 lists the structures of polyketides that were produced by marine-derived
Bacillus species.
Figure 6. Polyketides produced by marine-derived Bacillus species.
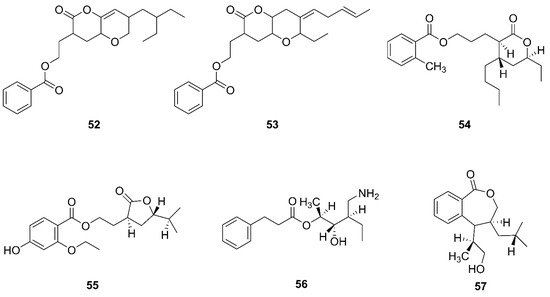
Two novel compounds, O-heterocycle pyrans, 2-(7-(2-Ethylbutyl)-2,3,4,4a,6,7-hexahydro-2-oxopyrano-[3,2b]-pyran-3-yl)-ethyl benzoate (
52) and 2-((4Z)-2-ethyl-octahydro-6-oxo-3-((E)-pent-3-enylidene)-pyrano-[3,2b]-pyran-7-yl)-ethyl benzoate (
53) were obtained by repeated chromatography from the heterotrophic bacterium
B. subtilis MTCC 10407 associated with brown seaweed
Sargassum myriocystum on the southeast coast of India
[61][64]. Additionally, 11-(15-butyl-13-ethyl-tetrahydro-12-oxo-2H-pyran-13-yl) propyl-2-methylbenzoate (
54), 9-(tetrahydro-2-isopropyl-11-oxofuran-10-yl)-ethyl-4-ethoxy-2-hydroxybenzoate (
55), 12-(aminomethyl)-11-hydroxyhexanyl-10-phenylpropanoate (
56), and 7-(14-hydroxypropan-13-yl)-8-isobutyl-7, and 8 dihydrobenzo[c]oxepin-1(3H)-one (
57) were isolated from a heterotrophic marine bacterium
B. amyloliquefaciens. This strain was isolated from the brown seaweed
Padina gymnospora collected from the intertidal zone of the Mannar Gulf in Peninsular India
[60][63].
7. Macrolactins
Marine
Bacillus species produce abundant polyketide classes of antibiotic agents, such as macrolactins, difficidins, and bacillaenes
[13][36][62][13,36,65]. Diffcidin is a highly unsaturated macrocyclic polyene with a 22-membered carbon skeleton and a phosphate moiety, which is rarely found in secondary metabolites of
Bacillus species. Bacillaene is a linear structure consisting of a conjugated hexaene. Carbon skeleton of most macrolactins contains three diene groups attached to the carbon backbone of a 24-membered lactone ring
[36].
Figure 7 lists the structures of macrolactins produced by marine-derived
Bacillus species.
Figure 7. Macrolactins produced by marine-derived Bacillus species.
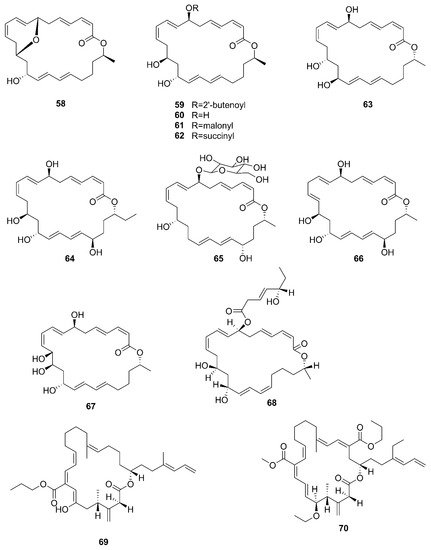
A new macrolactin derivative, 7,13-epoxyl-macrolactin A (
58), along with four known macrolactins, 7-O-2′E-butenoyl macrolactin A (
59), Macrolactin A (
60), 7-O-malonyl macrolactin A (
61), and 7-O-succinyl macrolactin A (
62) were isolated from bacteria
B. subtilis B5. It is worth emphasizing that this strain was extracted from deep-sea sediments at depths of 3000 m in the Pacific Ocean
[63][66]. Compounds
60 and
62 were also isolated from seaweed-associated
B. amyloliquefaciens MTCC 10456 in the MTCC of Chandigarh, India; this is the first report on the co-production and isolation of anti-
Malassezia spp. compounds from marine
Bacillus species
[41][44]. In particular, the major difference between compounds
58 and
59–
62 is in the epoxy ring. Compound
58 displayed a potent inhibitory effect on the expression of interleukin-1
β (IL-1
β), interleukin-6 (IL-6) and inducible nitric oxide synthase (iNOS), due to the existence of the epoxy ring
[63][66]. Five novel 24-membered macrolactins named bamemacrolactins A (
63), B (
64), C (
65), D (
66) and E (
67) were produced by
B. siamensis, which was isolated from the gorgonian coral
Anthogorgia caerulea gathered from Beihai city (Guangxi, China)
[64][67]. The 7-O-methyl-5′-hydroxy-3′-heptenoate−macrolactin (
68), a new macrolactin compound, was obtained from
B. subtilis MTCC10403 associated with seaweed
Anthophycus longifolius collected from the Gulf of Mannar of Peninsular India
[65][68]. Compounds
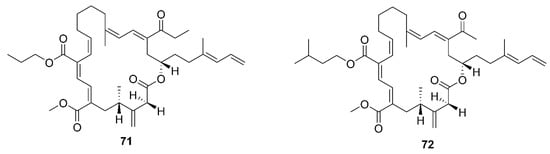
69–
72 were characterized as four homologous difficidin-type 21-membered macrocyclic lactone, isolated from a heterotrophoic
B. amyloliquefaciens MTCC12713 associated with an intertidal macroalga
Kappaphycus alverezii collected from the Gulf of Mannar in Peninsular India. In addition, they were established as 18,19-dihydro-6-hydroxy-8-propyl carboxylate difficidin, 5-ethoxy-28-methyl-(9-methyl-19propyl dicarboxylate) difficidin, (6-methyl-9-propyl dicarboxylate)-19-propanone difficidin, and 20-acetyl-(6-methyl-9-isopentyl dicarboxylate) difficidin, respectively
[13].
8. Other Compounds
Figure 8 lists the other compounds produced by marine-derived
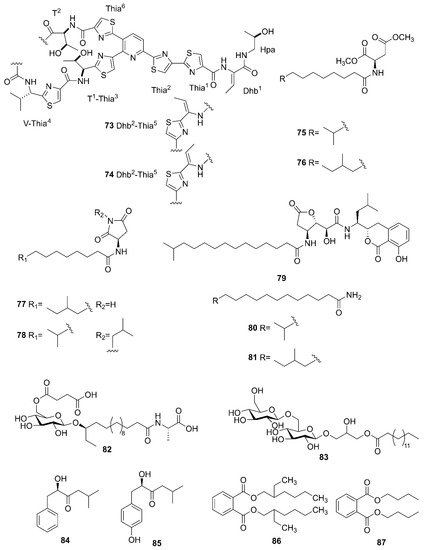
Bacillus species. A novel thiopeptide named micrococcin P3 (
73) and a known compound named micrococcin P1 (
74) were isolated from the fermentation broth of
B. stratosphericus [66][69]. Five new bacillamidins A (
75), B (
76), C (
77), D (
78) and E (
79), along with two known synthetic analogs, bacillamidins F (
80) and G (
81), were isolated from the marine-derived
B. pumilus strain RJA1515. This strain was extracted from deep-sea sediments at depths of 84 m collected in Bamfield in British Columbia
[67][70]. Ieodoglucomide C (
82) and ieodoglycolipid (
83), two new glycolipids, were produced by the marine-derived
B. licheniformis 09IDYM23 which was isolated from sediments at a depth 20 m collected at Ieodoin the southern reef of the Republic of Korea, both of which were obtained from the fermentation of this strain
[68][71]. According to bioactivity-guided strategy, (-)-sattabacin (
84) and (-)-4-hydroxysattabacin (
85) were firstly discovered from
Bacillus sp. (SCO-147) collected from marine sediments in Suncheon Bay of Korea
[69][72]. Marine-derived
B. subtilis AD35, gathered from marine water and sediment at the Alexandria sea shore in Egypt, could yield a previously reported but firstly isolated compound, Di-(2-ethylhexyl) phthalate (DEHP) (
86)
[70][73]. Additionally, compound
86 and dibutyl phthalate (DBP) (
87) were isolated from the extract broth of marine-derived
B. polymyxa L
1-9, which was collected in a mud sample from the intertidal mudflat in the Lianyungang Port of China
[71][74].
Figure 8. Other compounds produced by marine-derived Bacillus species.
AIn this review, a total of 87 secondary metabolites were reported from marine-derived
Bacillus species from January 2014 to December 2021. Their chemical structures were classified into cyclic lipopeptides (
1–
19, among them,
1–
2 surfactins,
3–
10 belong to iturins,
11 belongs to plipastatin), diketopiperazines (
20–
27), linear lipopeptides (
28–
38), nonribosomal peptides (
39–
51), polyketides (
52–
57), macrolactins (
58–
72, among them,
69–
72 belong to difficidins), and other compounds (
73–
87) according to their putative biogenetic sources. As shown in
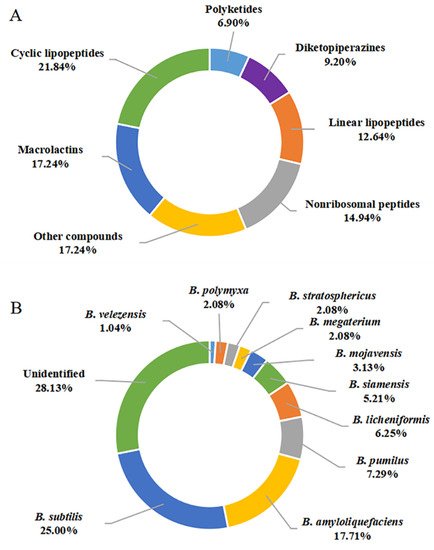
Figure 9A, 21.84% of the compounds reported were CLPs, and these compounds account for an overwhelming majority of all 87 metabolites, followed by macrolactins with 17.24%. Therefore, CLPs are a class of secondary metabolites with structural diversity and pharmacological perspective.
Figure 9. Quantification of studies on secondary metabolites from marine-derived
Quantification of this studies on secondary metabolites from marine-derived
Bacillus
: (
A
) Chemical structures categories; (
B
) Producing strains; (
C
) Environment sources.
The genus
Bacillus comprises more than 350 species, some of which are used as antifungal agents, while others are promising producers of green pesticide
[14]. As discussed above, a total of 10 identified species, including
B. subtilis,
B. amyloliquefaciens,
B. megaterium CGMCC7086,
B. mojavensis B0621A,
B. licheniformis,
B. siamensis,
B. stratosphericus,
B. pumilus RJA1515,
B. polymyxa L
1-9, and
B. velezensis SH-B74 were reported as the producing strains of these described secondary metabolites. Among them,
B. subtilis and
B. amyloliquefaciens were the most prolific strains, with 24 (20.00%) and 17 (17.71%) metabolites identified, respectively (
Figure 10B). CLPs are ubiquitous in several
Bacillus strains. However, linear lipopeptides are found predominantly in
B. subtilis species, while the macrolactins are more preponderant in
B. amyloliquefaciens species, suggesting the species-specific metabolites.
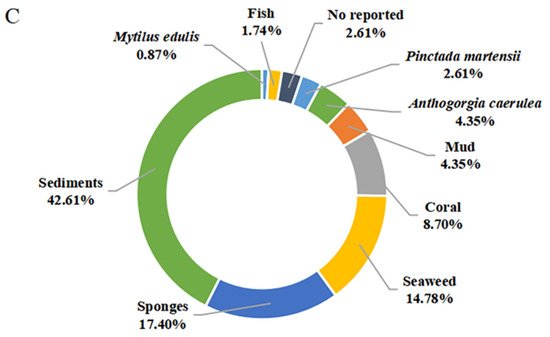
The secondary metabolites of marine-derived
Bacillus species could be isolated from marine sediments, marine invertebrates (sponges, molluscs, and corals), and vertebrates (mainly fishes), as well as marine plants (mainly seaweed). Currently, there are 9 reported sources of
Bacillus secondary metabolites. As shown in
Figure 9C, a total of 49 compounds {
1–
7,
11,
15–
18,
28–
37,
39–
48,
58–
62,
75–
85,
86 (
B. subtilis AD35)} originated from marine sediments, accounting for 42.61% of the sources of
Bacillus species. In particular, the producing strains of
58–
62 originated from deep-sea sediment at a depth of 3000 m, while the other strains of
75–
83 originated from deep-sea sediment at depths less than 100 m. Moreover, the producing strains of
12–
14, 86 (
B. polymyxa L
1-9), and
87 originated from mud. More precisely, the producing strains of
15–
16 and
28–
37 originated from the Republic of Korea’s southern reef. Twenty compounds (
20–
27,
39–
48,50,51), identified from marine sponges, accounted for 17.40% of the reported environmental sources of
Bacillus secondary metabolites. It is worth noting that
20 belonging to diketopiperazine were isolated unprecedentedly from a sponge-associated microbe. Moreover,
45–
49,
52–
56 and
63–
67 were also derived from coral, wherein
63–
67, whose producing strains were obtained from the gorgonian coral
A. caerulea, accounted for 4.35% of the reported environmental sources of
Bacillus secondary metabolites. The
8–
10 and
19 that originated from pearl oyster
P. martensii and
M. edulis, respectively, accounted for 3.48% of the reported ones. A total of 17 secondary metabolites {
38,45,
47,
49,
52–
57,
60,
62 (
B. amyloliquefaciens MTCC 10456) and
68–
72} were identified from
Bacillus residing in marine plants, accounted for 14.78% of the reported total amount. Thereinto, the producing strains of
52–
57 originated from brown seaweed, while the producing strains of
45,
47,
49,
60,
62 (
B. amyloliquefaciens MTCC 10456),
38,
52–
57 and
68–
72 were collected from seaweed. In addition to these producing strains, only one
Bacillus strain (
B. megaterium CGMCC7086), which produced
45 and
49, was obtained from the intestines of marine fish and accounted for 1.74% of the reported total amount. Unfortunately, the environmental sources of the producing strain of
47 (
B. subtilis B38),
73 and
74 (
B. stratosphericus) were not described. From the above analysis, it can be concluded that marine sediments and sponges are more abundant sources of productive strains of marine-derived
Bacillus, and which deserved much more attention in subsequent chemical studies.