Aegerolysins are remarkable proteins. They are distributed over the tree of life, being relatively widespread in fungi and bacteria, but also present in some insects, plants, protozoa, and viruses. Their function, in particular, is intriguing. Aegerolysin proteins are involved in various interactions by recognizing a molecular receptor in the target organism. Despite their abundance in cells of certain developmental stages and their presence in secretomes, only a few aegerolysins have been studied in detail. Formation of pores with various larger non-aegerolysin-like protein partners is one of the possible responses of the aegerolysin-producing organism in competitive exclusion of other organisms from the ecological niche.
- aegerolysins
- bacteria
- fungi
- insecticidal
- lipid binding
- lifestyle
- membrane-attack complex/perforin domain (MACPF)
- pore forming proteins
1. Introduction
2. Aegerolysins
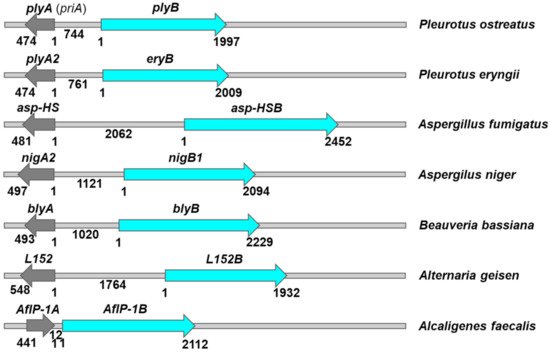
ResWearchers have collected (experimental) published data on 23 different aegerolysins and their variants. In total, they were characterized from 18 different species belonging to different kingdoms of tree of life. Twelve of these aegerolysins belong to fungi, four to bacteria, and one to insects and viruses. In fungi, they were characterized from four mushrooms (Agaricomycotina) from the order Agaricales: Pleurotus ostreatus, P. eryngii, Agrocybe aegerita, and Moniliophthora pernicious, as well as in the ordo Polyporales: Lignosus rhinocerotis (Table 1). The origin of these aegerolysins were also four filamentous Eurotimycetes from the ordo Eurotiales: Aspergillus fumigatus, A. niger, A. terreus, and A. oryzae (Table 1). Two species belonged to the Sordariomycetes, ordo Hypocreales: Beauveria bassiana, and Trichoderma atroviride, and another to the Dothideomycetes, ordo Pleosporales: Alternaria geisen (Table 1). There were four bacterial species, two belonging to Firmicutes: Bacillus thuringiensis and Clostridium bifermentans, and another two to Proteobacteria: Pseudomonas aeruginosa and Alcaligenes faecalis (Table 1). Another species belongs to Insecta—Lepidoptera, Noctuidae: Pseudoplusia includes, and another to Varidnaviria, Ascoviridae: Trichoplusia ni ascovirus 2c (Table 1).| Organism Name | Other Names | Taxonomy | Lifestyle/Niche | Reference | |||
|---|---|---|---|---|---|---|---|
| Fungi | |||||||
| Pleurotus ostreatus | Oyster mushroom Hiratake |
Agaricomycotina | Agaricales | Saprotroph White rot Nematocidal |
[12] | [15] |
| Short Name | Aegerolysin/ Structure |
Membrane Receptor |
Function | Partner Protein Short Name | Partner Protein/ Structure |
Organism | Reference | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| PlyA | Pleurotolysin A PDB ID: 4OEBA |
SM/Chol | n.d. | PlyB | Pleurotolysin B PDB ID: 4OEJ Membrane embedded PlyA/PlyB pore PDB ID: 4V2T |
Pleurotus ostreatus | [56][57] | [59,61] | ||||||||||||
| Pleurotus eryngii | King oyster or trumpet or brown mushroom Boletus of the steppes | |||||||||||||||||||
| OlyA | French horn mushroom Aliʻi oyster |
Agaricomycotina | Agaricales | Ostreolysin A | SM/Chol CPE/Chol Lipid raftsSaprotroph Grassland-litter decomposer Facultatively biotrophic Nematocidal |
[ | Involvement in mushroom fruiting Anticancer (+PlyB) | 13] | [16] | |||||||||||
| PlyB | Pleurotolysin B | PDB ID: | 4OEJ | Pleurotus ostreatus | [58][59][60][61][62][63][64][65][66][67][68][69][70][71][72][73] | [63,64 | Cyclocybe aegerita | Agrocybe aegerita | Poplar mushroom Tea tree mushroom Cha shu gu Yanagi-matsutake Sword-belt mushroom Velvet pioppini |
Agaricomycotina | Agaricales | Saprotroph Weak white rot on hardwoods Facultatively pathogenic |
[14] | [17] | ||||||
| ] | [ | 80 | ] | [ | 81 | ] | [82] | [72,73,74,75,78,79,80,81,82,83,87] | Moniliophthora perniciosa | Crinipellis perniciosa | Witches’ broom disease |
Agaricomycotina | Agaricales | Hemibiotrophic plant pathogen Broad range of host |
||||||
| rOly | Recombinant ostreolysin | Lipid rafts? | Antiproliferative Pro-apoptotic | [15] | [18] | |||||||||||||||
| n.d. | n.d. | Pleurotus ostreatus | [ | 83 | ] | [ | 84][85] | [88,89,90] | Lignosus rhinocerotis | |||||||||||
| PlyA2 | Tiger milk mushroom | Agaricomycotina | Polyporales | Pe.PlyA/Pleurotolysin A2Saprotroph | SM/Chol CPE/Chol CPE White rot |
Lipid rafts | [16] | [19] | ||||||||||||
| Insecticidal | (+PlyB) | EryB | Erylysin B | Pleurotus eryngii | [ | 12][68][78][86][87][88][89] | [15,79,80,93,94,95,96] | Neosartorya fumigata | Aspergillus fumigatus | Eurotimycetes | Eurotiales | Saprotroph Ubiquitous in soil and compost Human (opportunistic) pathogen |
[17][18][19][20] | [20,21,22,23] | ||||||
| EryA | Erylysin A | CPE/Chol CL/DPPC/Chol |
Insecticidal (+PlyB) Inhibition of cytokinesis |
No | No | Pleurotus eryngii | [53][68][89][90][91] | [78,80,96,98,99] | Aspergillus niger | Eurotimycetes | Eurotiales | Saprotroph Ubiquitous in soil and compost Human opportunistic pathogen |
[ | |||||||
| Aa-Pri1 | Aegerolysin Aa-Pri1 | 17 | n.d. | ][18][21] | [22][23] | [20 | [ | ,21 | 24 | ,24,25 | ] | ,26,27] | ||||||||
| No | No | Agrocybe aegerita | [ | 58 | ] | [ | 92] | [63,100] | Aspergillus terreus | Eurotimycetes | Eurotiales | Saprotroph Human opportunistic pathogen |
[17][18][25] | [ | ||||||
| MpPRIA1 | 20 | , | 21 | ,28] | ||||||||||||||||
| Putative aegerolysin | n.d. | MpPLYB | ? | n.d. | Moniliophthora perniciosa | [93] | [101] | |||||||||||||
| MpPRIA2 | Putative aegerolysin | n.d. | MpPLYB | ? | n.d. | Moniliophthora perniciosa | [93] | [101] | 30][31][32] | [32,33,34,35] | ||||||||||
| Alternaria geisen | Black spot of Japanese pear | Dothideomycetes | Pleosporales | Plant pathogen | [33][34] | [36,37] | ||||||||||||||
| Bacteria | ||||||||||||||||||||
| Bacillus thuringiensis | Firmicutes | Ubiquitous opportunistic pathogen on vertebrates, plants, insects, nematodes, mollusks, protozoan, animal, and human parasites. Soils, grain dusts, dead insects, water Aerobic and spore-forming |
[35][36] | [38,39] | ||||||||||||||||
| , | 65 | , | 66 | , | 67 | , | 68,69,70,71,76,80,84,85,86,102,104] | |||||||||||||
| OlyA6 | Ostreolysin A6 PDB ID: 6MYJ |
SM/Chol CPE/Chol CAEP/POPC/Chol Lipid rafts |
Insecticidal (+PlyB) | PlyB | Pleurotolysin B PDB ID: 4OEJ |
Pleurotus ostreatus | [53][68][74][75][76][77][78][79 | Aspergillus oryzae | Eurotimycetes | Eurotiales | Saprotroph | [17][18][26] | [20,21,29] | |||||||
| Beauveria bassiana | Sordariomycetes | Hypocreales | Entomopathogen Endophyte Soil and insects |
[27 | ||||||||||||||||
| GME7309 | ] | [ | 28 | ] | [30, | GME7309_g aegerolysin-domain-containing protein31] | ||||||||||||||
| n.d. | n.d. | n.d. | Lignosus rhinocerotis | [ | 94 | ] | [105] | Hypocrea atroviridis | Trichoderma atroviride | Sordariomycetes | Hypocreales | Mycoparasitic (including oomycetes) Cosmopolitan, soil |
[29 | |||||||
| Asp-HS | Asp-hemolysin | ] | Oxidized low-density lipoproteins | Cytotoxic effects on murine macrophages and vascular endothelial cells Induce cytokine genes | [ | Asp-HSB | n.d. | Aspergillus fumigatus | [95][96][97][98][99][100]] | [107,108 | [ | ,109 | 101] | ,110 | [102] | ,111 | [ | ,112 | 103 | ,113,114,115] |
| Asp-HS-like | Asp hemolysin-like | n.d. | n.d. | No | No | Aspergillus | fumigatus | [102] | [114] | |||||||||||
| NigA1 | Nigerolysin A1 | n.d. | n.d. | No | No | Aspergillus niger | [104][105] | [91,92] | ||||||||||||
| NigA2 | Nigerolysin A2 | CPE/Chol | n.d. | NigB1 | Nigerolysin B1 | Aspergillus niger | [104][105] | [91,92 | Paraclostridium bifermentans | subsp. malaysia | Clostridium bifermentans | subsp. malaysia | Firmicutes | Anaerobic, forming endospores Mosquito larvicidal |
[37] | [40] | ||||
| ] | ||||||||||||||||||||
| Ter | Terrelysin | n.d. | n.d. | No | No | Aspergillus terreus | [25][106][107] | [28,116,117] | Alcaligenes faecalis | Proteobacteria | Soil, water, environments associated with humans Human opportunistic pathogen Nematocidal |
[38 | ||||||||
| AoHlyA | Aspergillus oryzae | hemolysin | n.d. | ] | [ | n.d.41] | ||||||||||||||
| No | No | Aspergillus oryzae | [ | 108 | ] | [ | 109][110] | [118,119,120] | Pseudomonas aeruginosa | Proteobacteria | Ubiquitous opportunistic pathogen on: | |||||||||
| BlyA | humans, vertebrates, plants, and insects | Beauveriolysin A | [ | SM/Chol CPE/Chol | 39 | n.d. | ] | BlyB[40][41][42][43] | [42,43,44,45,46] | |||||||||||
| Beauveriolysin B | Beauveria bassiana | [ | 54 | ] | [ | 52 | ] | Insecta | ||||||||||||
| Agl1 | Trichoderma atroviride | aegerolysin | Conidiation Antagonism |
n.d. | No | No | Trichoderma atroviride | [111] | [128] | Chrysodeixis includens | Pseudoplusia includes | Lepidoptera | Noctuidae | Plant pest (defoliator) Larvae feed on a wide range of plants |
[ | |||||
| L152 | Alternaria geisen | aegerolysin | n.d. | 44][45] | [47 | n.d.,48] | ||||||||||||||
| L152B | n.d. | Alternaria geisen | Viria | |||||||||||||||||
| Trichoplusia ni | ascovirus 6a1 | Trichoplusia ni | ascovirus 2c | Varidnaviria | Ascoviridae | Obligate pathogen | Pseudoplusia includens | moth larvae | [44][46][47][48] | [47,49,50,51] |
3. Aegerolysin Binary Partner Proteins
| [ | ||||||||||||||||||
| 33 | ||||||||||||||||||
| ] | ||||||||||||||||||
| [ | ||||||||||||||||||
| 36 | ||||||||||||||||||
| ] | ||||||||||||||||||
| Cry34Ab1 | ||||||||||||||||||
| (Gpp34Ab1) | ||||||||||||||||||
| 13.6 kDa Insecticidal crystal protein | ||||||||||||||||||
| PDB ID: 4JOX | ||||||||||||||||||
| Unknown protein receptor | ||||||||||||||||||
| Insecticidal | ||||||||||||||||||
| (+Cry34Ab1) | Cry35Ab1 | (Tpp35Ab1) | 43.8 kDa insecticidal crystal protein PDB ID: 4JP0 |
Bacillus thuringiensis | [36][49] | 129 | [50][112][113][114][115] | ,130 | [ | ,131 | 116 | ,132 | ][ | ,133 | 117] | [39,,134,135,136] | ||
| Cbm17.1 | Hemolysin-like protein Cbm17.1 | n.d. | Insecticidal (+Cry16Aa/Cry17Aa/ Cbm17.2) |
Cry16Aa, Cry17Aa, Cbm17.2 | Pesticidal crystal-like protein Cry16Aa and Cry17Aa, Hemolysin-like protein Cbm17.2 | Clostridium bifermentans | [37][118][119] | [40,140,142] | ||||||||||
| Cbm17.2 | Hemolysin-like protein Cbm17.2 | n.d. | Insecticidal (+Cry16Aa/Cry17Aa/ Cbm17.1) |
Cry16Aa, Cry17Aa, Cbm17.2 | Pesticidal crystal-like protein Cry16Aa and Cry17Aa, Hemolysin-like protein Cbm17.1 | Clostridium bifermentans | [37][118][119] | [40,140,142] | ||||||||||
| AfIP-1A | Two-component insecticidal protein 16 kDa unit PDB ID: 5V3S |
Unknown protein receptor AfIP-1A/AfIP-1B membrane pore |
n.d. | AfIP-1B | Two-component insecticidal protein 77 kDa unit |
Alcaligenes faecalis | [51][120] | [143,144] | ||||||||||
| RahU | RahU protein PDB ID: 6ZC1 |
CPE/Chol | n.d. | No | No | Pseudomonas aeruginosa | [121][122] | [145,148] | ||||||||||
| P23 | n.d. | n.d. | n.d. | No | No | Pseudoplusia includes | [45] | [48] | ||||||||||
| TnAV2cgp029 | n.d. | n.d. | n.d. | No | No | Trichoplusia ni ascovirus 2c | [46][47][123][124][125] | [49,50,149,150,152] |