Advancements in the clinical practice of non-small cell lung cancer (NSCLC) are shifting treatment paradigms towards increasingly personalized approaches. Liquid biopsies using various circulating analytes provide minimally invasive methods of sampling the molecular content within tumor cells. Plasma-derived circulating tumor DNA (ctDNA), the tumor-derived component of cell-free DNA (cfDNA), is the most extensively studied analyte and has a growing list of applications in the clinical management of NSCLC. As an alternative to tumor genotyping, the assessment of oncogenic driver alterations by ctDNA has become an accepted companion diagnostic via both single-gene polymerase chain reactions (PCR) and next-generation sequencing (NGS) for advanced NSCLC. ctDNA technologies have also shown the ability to detect the emerging mechanisms of acquired resistance that evolve after targeted therapy. Furthermore, the detection of minimal residual disease (MRD) by ctDNA for patients with NSCLC after curative-intent treatment may serve as a prognostic and potentially predictive biomarker for recurrence and response to therapy, respectively. Finally, ctDNA analysis via mutational, methylation, and/or fragmentation multi-omic profiling offers the potential for improving early lung cancer detection.
- Early detection
- Lung cancer
- NLST
- LDCT
- Pulmonary nodules
- DELFI
- Lung-CLiP
- CCGA
- non-small cell lung cancer
- NSCLC
1. Introduction
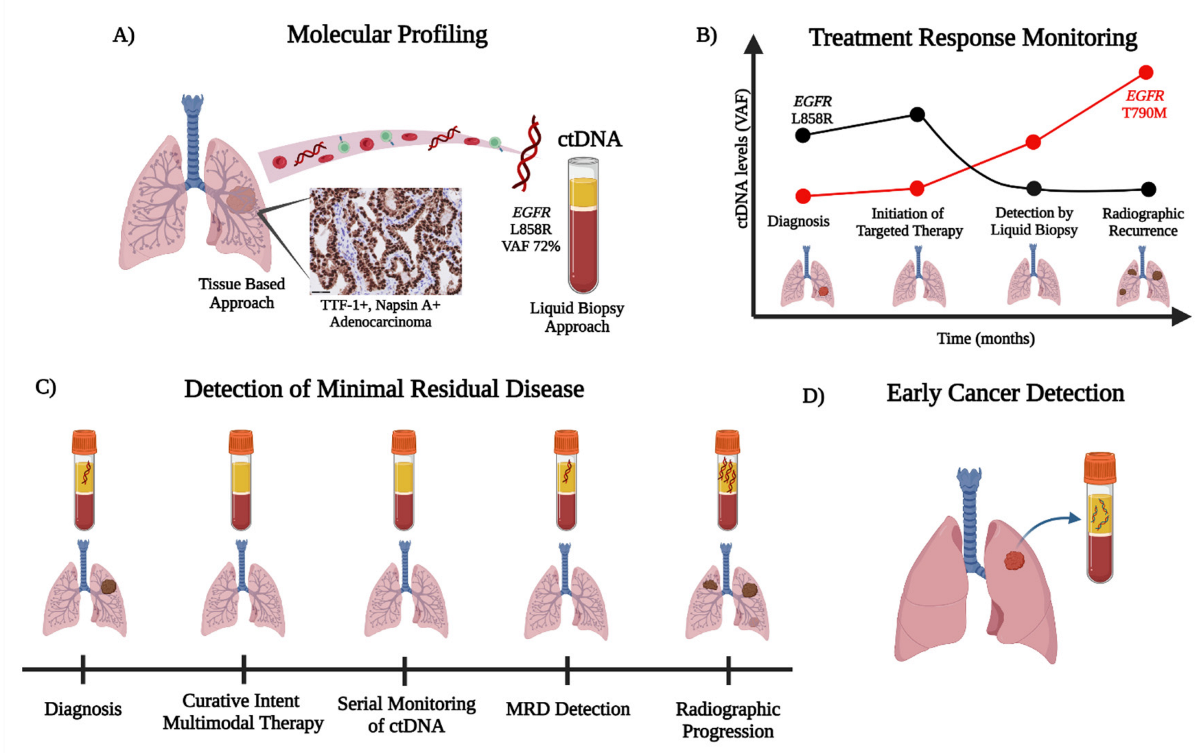
2. ctDNA for Early Cancer Detection
3. Conclusion
References
- Leighl, N.B.; Page, R.D.; Raymond, V.M.; Daniel, D.B.; Divers, S.G.; Reckamp, K.L.; Villalona-Calero, M.A.; Dix, D.; Odegaard, J.I.; Lanman, R.B.; et al. Clinical Utility of Comprehensive Cell-free DNA Analysis to Identify Genomic Biomarkers in Patients with Newly Diagnosed Metastatic Non–small Cell Lung Cancer. Clin. Cancer Res. 2019, 25, 4691–4700.
- Aggarwal, C.; Thompson, J.C.; Black, T.A.; Katz, S.I.; Fan, R.; Yee, S.S.; Chien, A.; Evans, T.L.; Bauml, J.M.; Alley, E.W.; et al. Clinical Implications of Plasma-Based Genotyping With the Delivery of Personalized Therapy in Metastatic Non–Small Cell Lung Cancer. JAMA Oncol. 2019, 5, 173–180.
- Rolfo, C.; Mack, P.; Scagliotti, G.V.; Aggarwal, C.; Arcila, M.E.; Barlesi, F.; Bivona, T.; Diehn, M.; Dive, C.; Dziadziuszko, R.; et al. Liquid Biopsy for Advanced NSCLC: A Consensus Statement From the International Association for the Study of Lung Cancer. J. Thorac. Oncol. 2021, 16, 1647–1662.
- Forshew, T.; Murtaza, M.; Parkinson, C.; Gale, D.; Tsui, D.W.Y.; Kaper, F.; Dawson, S.-J.; Piskorz, A.M.; Jimenez-Linan, M.; Bentley, D.; et al. Noninvasive Identification and Monitoring of Cancer Mutations by Targeted Deep Sequencing of Plasma DNA. Sci. Transl. Med. 2012, 4, 136ra68.
- Miller, A.; Shah, R.; Pentsova, E.I.; Pourmaleki, M.; Briggs, S.; Distefano, N.; Zheng, Y.; Skakodub, A.; Mehta, S.A.; Campos, C.; et al. Tracking tumour evolution in glioma through liquid biopsies of cerebrospinal fluid. Nature 2019, 565, 654–658.
- Crowley, E.; Di Nicolantonio, F.; Loupakis, F.; Bardelli, A. Liquid biopsy: Monitoring cancer-genetics in the blood. Nat. Rev. Clin. Oncol. 2013, 10, 472–484.
- Diaz, L.A., Jr.; Bardelli, A. Liquid Biopsies: Genotyping Circulating Tumor DNA. J. Clin. Oncol. 2014, 32, 579–586.
- Zhu, L.; Sun, H.-T.; Wang, S.; Huang, S.-L.; Zheng, Y.; Wang, C.-Q.; Hu, B.-Y.; Qin, W.; Zou, T.-T.; Fu, Y.; et al. Isolation and characterization of exosomes for cancer research. J. Hematol. Oncol. 2020, 13, 152.
- Alix-Panabières, C.; Pantel, K. Liquid Biopsy: From Discovery to Clinical Application. Cancer Discov. 2021, 11, 858–873.
- Nuzzo, P.V.; Berchuck, J.E.; Korthauer, K.; Spisak, S.; Nassar, A.H.; Alaiwi, S.A.; Chakravarthy, A.; Shen, S.Y.; Bakouny, Z.; Boccardo, F.; et al. Detection of renal cell carcinoma using plasma and urine cell-free DNA methylomes. Nat. Med. 2020, 26, 1041–1043.
- Best, M.G.; Wesseling, P.; Wurdinger, T. Tumor-Educated Platelets as a Noninvasive Biomarker Source for Cancer Detection and Progression Monitoring. Cancer Res. 2018, 78, 3407–3412.
- Mandel, P.; Metais, P. Nuclear Acids In Human Blood Plasma. Comptes Rendus Seances Soc. Biol. Ses Fil. 1948, 142, 241–243.
- Corcoran, R.B.; Chabner, B.A. Application of Cell-free DNA Analysis to Cancer Treatment. N. Engl. J. Med. 2018, 379, 1754–1765.
- Jahr, S.; Hentze, H.; Englisch, S.; Hardt, D.; Fackelmayer, F.O.; Hesch, R.D.; Knippers, R. DNA fragments in the blood plasma of cancer patients: Quantitations and evidence for their origin from apoptotic and necrotic cells. Cancer Res. 2001, 61, 1659–1665.
- Wan, J.C.M.; Massie, C.; Garcia-Corbacho, J.; Mouliere, F.; Brenton, J.D.; Caldas, C.; Pacey, S.; Baird, R.; Rosenfeld, N. Liquid biopsies come of age: Towards implementation of circulating tumour DNA. Nat. Rev. Cancer 2017, 17, 223–238.
- Pisetsky, D.S.; Fairhurst, A.-M. The origin of extracellular DNA during the clearance of dead and dying cells. Autoimmunity 2007, 40, 281–284.
- El Messaoudi, S.; Rolet, F.; Mouliere, F.; Thierry, A.R. Circulating cell free DNA: Preanalytical considerations. Clin. Chim. Acta 2013, 424, 222–230.
- Kustanovich, A.; Schwartz, R.; Peretz, T.; Grinshpun, A. Life and death of circulating cell-free DNA. Cancer Biol. Ther. 2019, 20, 1057–1067.
- Thijssen, M.A.; Swinkels, D.W.; Ruers, T.J.M.; De Kok, J.B. Difference between free circulating plasma and serum DNA in patients with colorectal liver metastases. Anticancer Res. 2002, 22, 421–425.
- Umetani, N.; Hiramatsu, S.; Hoon, D.S. Higher Amount of Free Circulating DNA in Serum than in Plasma Is Not Mainly Caused by Contaminated Extraneous DNA during Separation. Ann. N. Y. Acad. Sci. 2006, 1075, 299–307.
- Siravegna, G.; Marsoni, S.; Siena, S.; Bardelli, A. Integrating liquid biopsies into the management of cancer. Nat. Rev. Clin. Oncol. 2017, 14, 531–548.
- Bettegowda, C.; Sausen, M.; Leary, R.J.; Kinde, I.; Wang, Y.; Agrawal, N.; Bartlett, B.R.; Wang, H.; Luber, B.; Alani, R.M.; et al. Detection of circulating tumor DNA in early- and late-stage human malignancies. Sci. Transl. Med. 2014, 6, 224.
- Pellini, B.; Szymanski, J.; Chin, R.-I.; Jones, P.A.; Chaudhuri, A.A. Liquid Biopsies Using Circulating Tumor DNA in Non-Small Cell Lung Cancer. Thorac. Surg. Clin. 2020, 30, 165–177.
- Lim, C.; Tsao, M.S.; Le, L.W.; Shepherd, F.A.; Feld, R.; Burkes, R.L.; Liu, G.; Kamel-Reid, S.; Hwang, D.; Tanguay, J.; et al. Biomarker testing and time to treatment decision in patients with advanced nonsmall-cell lung cancer. Ann. Oncol. 2015, 26, 1415–1421.
- U.S. Food and Drug Administration. FDA Approves First Liquid Biopsy Next-Generation Sequencing Companion Diagnostic Test. 2020. Available online: https://www.fda.gov/news-events/press-announcements/fda-approves-first-liquid-biopsy-next-generation-sequencing-companion-diagnostic-test (accessed on 23 March 2022).
- U.S. Food and Drug Administration. FDA Approves Liquid Biopsy NGS Companion Diagnostic Test for Multiple Cancers and Biomarkers. 2020. Available online: https://www.fda.gov/drugs/resources-information-approved-drugs/fda-approves-liquid-biopsy-ngs-companion-diagnostic-test-multiple-cancers-and-biomarkers (accessed on 23 March 2022).
- Adjuvant Durvalumab for Early Stage NSCLC Patients with ctDNA Minimal Residual Disease. Available online: https://clinicaltrials.gov/ct2/show/NCT04585477?term=NCT04585477&draw=2&rank=1 (accessed on 12 June 2022).
- Personalized Escalation of Consolidation Treatment Following Chemoradiotherapy and Immunotherapy in Stage III NSCLC in Stage III NSCLC. Available online: https://clinicaltrials.gov/ct2/show/NCT04585490?term=NCT04585490&draw=2&rank=1 (accessed on 12 June 2022).
- Kinde, I.; Wu, J.; Papadopoulos, N.; Kinzler, K.W.; Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 2011, 108, 9530–9535.
- National Lung Screening Trial Research Team; Aberle, D.R.; Adams, A.M.; Berg, C.D.; Black, W.C.; Clapp, J.D.; Fagerstrom, R.M.; Gareen, I.F.; Gatsonis, C.; Marcus, P.M.; et al. Reduced Lung-Cancer Mortality with Low-Dose Computed Tomographic Screening. N. Engl. J. Med. 2011, 365, 395–409.
- Welch, J.S. Use of Whole-Genome Sequencing to Diagnose a Cryptic Fusion Oncogene. JAMA 2011, 305, 1577–1584.
- Shen, S.Y.; Burgener, J.M.; Bratman, S.V.; De Carvalho, D.D. Preparation of cfMeDIP-seq libraries for methylome profiling of plasma cell-free DNA. Nat. Protoc. 2019, 14, 2749–2780.
- Chabon, J.J.; Hamilton, E.G.; Kurtz, D.M.; Esfahani, M.S.; Moding, E.J.; Stehr, H.; Schroers-Martin, J.; Nabet, B.Y.; Chen, B.; Chaudhuri, A.A.; et al. Integrating genomic features for non-invasive early lung cancer detection. Nature 2020, 580, 245–251.
- Abbosh, C.; Birkbak, N.; Swanton, C. Early stage NSCLC—Challenges to implementing ctDNA-based screening and MRD detection. Nat. Rev. Clin. Oncol. 2018, 15, 577–586.
- Hong, Y.; Kim, W.J. DNA methylation markers in lung cancer. Curr. Genom. 2021, 21, 79–87.
- Huang, J.; Soupir, A.C.; Schlick, B.D.; Teng, M.; Sahin, I.H.; Permuth, J.B.; Siegel, E.M.; Manley, B.J.; Pellini, B.; Wang, L. Cancer Detection and Classification by CpG Island Hypermethylation Signatures in Plasma Cell-Free DNA. Cancers 2021, 13, 5611.
- Qi, J.; Hong, B.; Tao, R.; Sun, R.; Zhang, H.; Zhang, X.; Ji, J.; Wang, S.; Liu, Y.; Deng, Q.; et al. Prediction model for malignant pulmonary nodules based on cfMeDIP-seq and machine learning. Cancer Sci. 2021, 112, 3918–3923.
- Klein, E.; Richards, D.; Cohn, A.; Tummala, M.; Lapham, R.; Cosgrove, D.; Chung, G.; Clement, J.; Gao, J.; Hunkapiller, N.; et al. Clinical validation of a targeted methylation-based multi-cancer early detection test using an independent validation set. Ann. Oncol. 2021, 32, 1167–1177.
- Liu, M.C.; Oxnard, G.R.; Klein, E.A.; Swanton, C.; Seiden, M.V. Sensitive and specific multi-cancer detection and localization using methylation signatures in cell-free DNA. Ann. Oncol. 2020, 31, 745–759.
- The SUMMIT Study: A Cancer Screening Study (SUMMIT). Available online: https://clinicaltrials.gov/ct2/show/NCT03934866?term=NCT03934866&draw=2&rank=1 (accessed on 12 June 2022).
- Mathios, D.; Johansen, J.S.; Cristiano, S.; Medina, J.E.; Phallen, J.; Larsen, K.R.; Bruhm, D.C.; Niknafs, N.; Ferreira, L.; Adleff, V.; et al. Detection and characterization of lung cancer using cell-free DNA fragmentomes. Nat. Commun. 2021, 12, 5060.
- Przybyl, J.; Chabon, J.J.; Spans, L.; Ganjoo, K.N.; Vennam, S.; Newman, A.M.; Forgó, E.; Varma, S.; Zhu, S.; Debiec-Rychter, M.; et al. Combination Approach for Detecting Different Types of Alterations in Circulating Tumor DNA in Leiomyosarcoma. Clin. Cancer Res. 2018, 24, 2688–2699.
