You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Jessie Wu and Version 1 by Jose Diaz-Chavez.
DNA methylation is an epigenetic mark that living beings have used in different environments. The MTases family catalyzes DNA methylation. This process is conserved from archaea to eukaryotes, from fertilization to every stage of development, and from the early stages of cancer to metastasis. The family of DNMTs has been classified into DNMT1, DNMT2, and DNMT3. Each DNMT has been duplicated or deleted, having consequences on DNMT structure and cellular function, resulting in a conserved evolutionary reaction of DNA methylation. DNMTs are conserved in the five kingdoms of life: bacteria, protists, fungi, plants, and animals.
- DNA methylation
- MTases
- RNAs
1. The Structure of MTAses
1. The Structure of DNA Methyltransferases
DNA methyltransferases (MTases) are conserved in living beings acting in orchestra with other epigenetic players. They have positioned themselves with a few exceptions as the main transcription regulators. MTases are a group with methyltransferase activity; they have evolved in different orthologs, but all have the methyltransferase domain and a DNA target recognition domain. Living beings have conserved MTases to survive the different and dynamic ambient conditions [2,3,4,5,19][1][2][3][4][5].
The reaction mechanism of MTases catalyzes DNA methylation in adenine or cytosine bases is known. All MTases interact with the cofactor S-adenosyl methionine (AdoMet) to transfer a methyl group and produce S-adenosyl-l-homocysteine (AdoHcy) and methylated DNA [20][6]. Moreover, another characteristic of all MTases is that they have three protein domains (in the carboxy-terminal domain for DNMTs): The adoMet binding domain, which interacts with AdoMet to obtain the methyl group; a target recognition domain (TRD), which recognize a short sequence of DNA to be targeted for methylation, and the catalytic domain, which transfers a methyl group to AdoMet to the targeted nucleotide [20][6] (Figure 1). These domains are the set of MTases, which beings have conserved in almost all species of living beings.
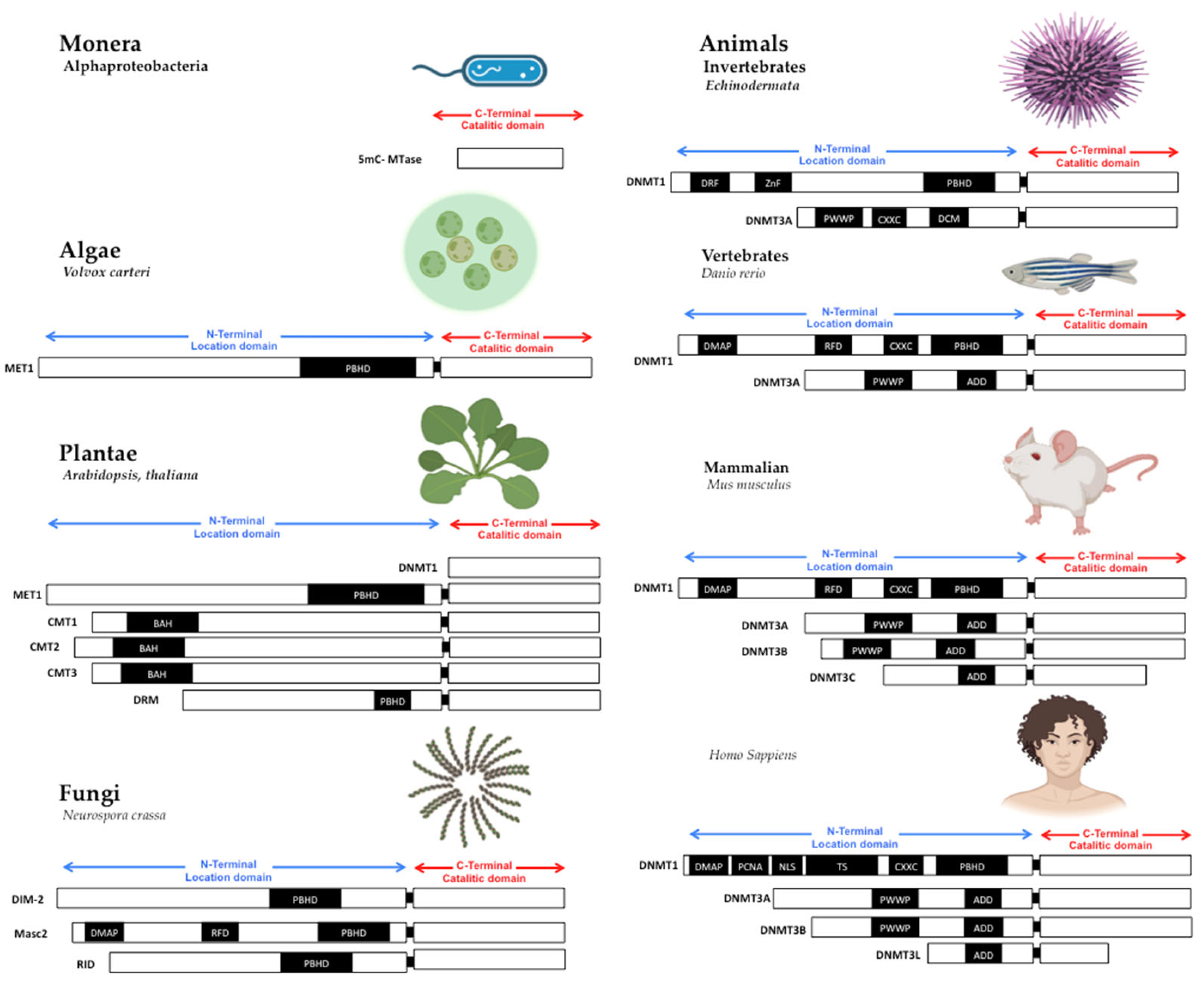
Figure 1. Structure of DNA methyltransferases. The DNMTs family is the DNA methylator in living beings. DNMTs have two domains: catalytic domain in the carboxy-terminal extreme, with the conserved catalytic motives, and location domain in the amino-terminal extreme, with the location and interaction chromatin motives. Monera has the catalytic domain mtase, Alphaproteobacteria. Protists and Algae are composed of MET1, CMT3, and DRM2 protein paralogs; MET1 is an example present in Volvox carteri. Fungi are composed of the protein paralogs: DIM-2, Masc1 and 2, and RID; the example is Neurospora crassa. In animals, Invertebrates, the protein paralogs are DNMT1 and DNMT3, in Echinoderma; Vertebrates, Fishes, DNMT1, and 3, zebrafish has 8 DNMT3 mammalians, the protein paralogs are: DNMT1 and DNMT3A/B/C in Mus musculus, and DNMT1, 3A/B/L in Homo sapiens. Abbreviatures: CD (chromo dominio), DMAP-1, binding domain (DMAP); motif to interact with PCNA (PCDNA); Nuclear localization Signal (NLS); Targeted Site (TS); Motif to Cys-X-X-Cys amino-acids, with zinc fingers (CXXC), Protein Binding Homeo Domain (PBHD); the motif of interaction with pro-trp-trp-pro (PWWP), and ATRX, DNMT3, DNMT3L domain (ADD). Note: Created with Biorender.com, accessed on 21 July 2022.
The primary sequence of MTases is essential to methyltransferase activity, but the shape is also crucial. The structural conformation domains and different motifs of MTases have been reviewed in other works [6[7][8],7], where domain or domains conformed MTases in all cases. First, in the amino-terminal domain, some motifs interact with CpG sequences as the CXXC domain in DNMT1 [21,22,23][9][10][11]. Still, without the CXXC motif, DNMT1 can’t actively interact with the PCNA motive to serve in DNA replication [24,25][12][13]; the PWWP motif in DNMT3A and DNMT3B, which interacts with chromatin proteins and localizes them in centromeric and pericentromeric chromatin [11,26][14][15]. The shape and the amino acid primary sequences of MTases are essential to be conserved in the catalytic domain in all living beings. The variable part on TRD is also evolving to give specificity to DNMTs duplications [25][13].
2. DNA Methyltransferases Are Regulated by Chemical Compounds and ncRNAs
The dysregulation of DNA methylation has a role in the development of cancer cells and other diseases [7,124][8][16]. As DNA methylation is a critical factor in global epigenetic regulation [125][17], it is not surprising to find DNMTs dysregulated in cancer. Actually, one of the main epigenetic characteristics in cancer is the global demethylation and local hypermethylation of the DNA [126][18]. There are multiple examples of how different tumors have either a misregulation of one or more DNMTs or even mutations. For example, hematological diseases like acute myeloid leukemia (AML) have mutations in the DNMT3A gene [127][19], whereas inactivating mutations in DNMT1 are related to genome-wide alterations of DNA methylation in colon cancer [128][20] (Figure 2). Different regulators of DNMTs have been researched, such as non-coding RNAs (ncRNA) and artificial compounds tested to influence DNA methylation. Targeting DNMTs is a promising tool to use alone or in combination to treat cancer. However, further research needs to be done in this field.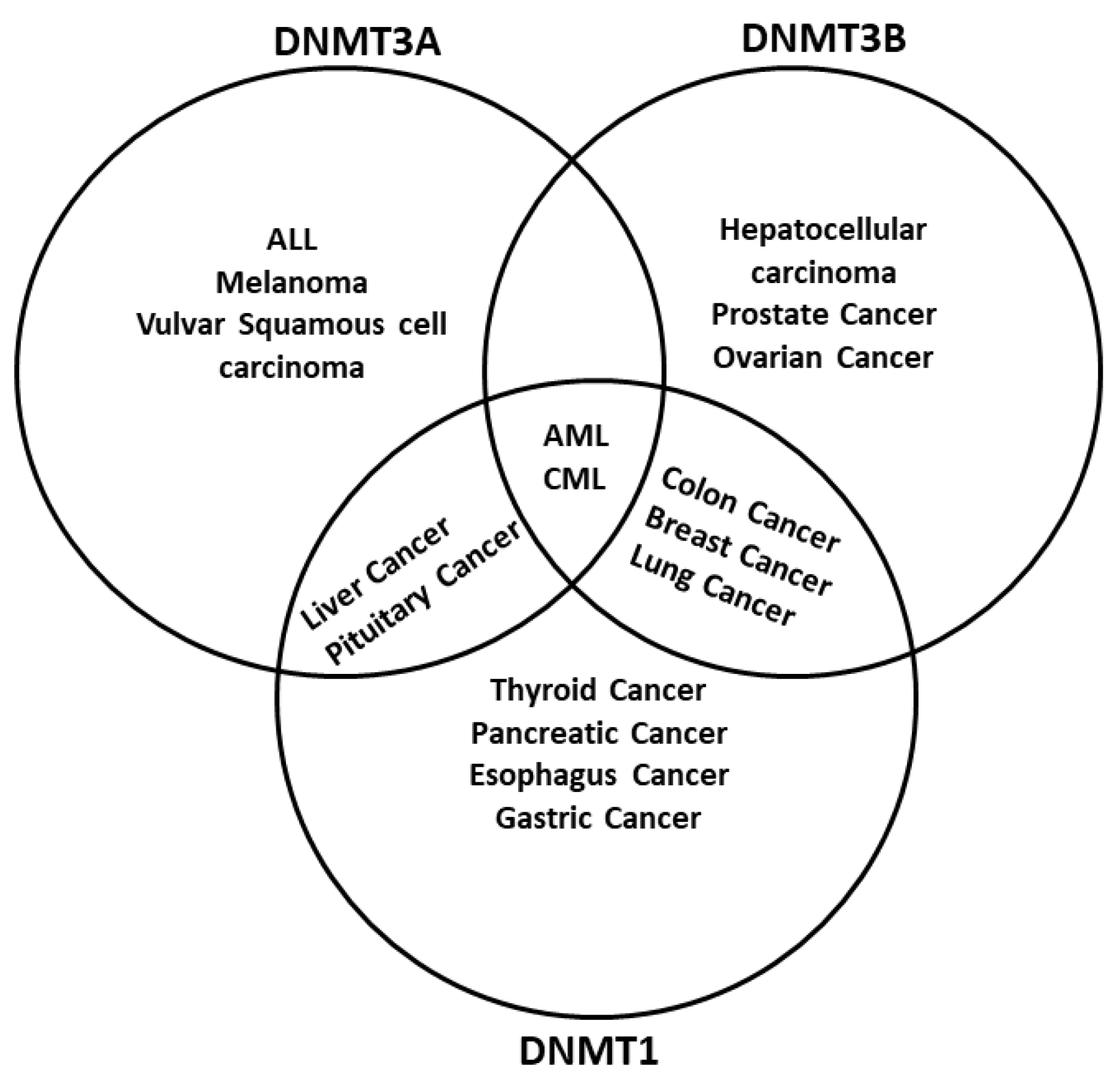
Figure 2. DNA methyltransferases are altered in cancer. DNMTs have a role in genomic regulation. In cancer, DNMTs are affected in expression level. In acute myeloid leukemia (AML) and chronic myeloid leukemia (CML), The three DNMTs are overexpressed. DNMT1 and DNMT3A have been described as affected in liver cancer and pituitary cancer, while DNMT1 and DNMT3B are overexpressed in breast cancer, colon cancer, and lung cancer; DNMT3B is deregulated in colon cancer and prostate cancer, and DNMT1 is deregulated in the pancreas cancer and esophagus cancer. In other cases, only one DNMT is overexpressed; however, only one DNMT could be enough to result in cancer development, progression, and metastasis. Abbreviatures: Myelodysplastic syndrome (MDS), chronic myelomonocytic leukemia (CMML), acute lymphoblastic leukemia (ALL), melanoma.
Table 1.
Studies of DNA methyltransferases (DNMTs) and non-coding RNAs in cancer.
| ncRNAs | DNMT Deregulated in Cancer |
Type of Cancer | Type of Deregulation | Reference |
|---|---|---|---|---|
| DBCCR1-003 | DNMT1 | Bladder | Down | [145][35] |
| linc-POU3F3 | DNMT1, 3A, and 3B | ESCC | Up | [149][36] |
| miR-148a | DNMT1 | Gastric | Down | [150][37] |
| miR-29a | DNMT1 | Liver | Down | [151][38] |
| miR-152 | DNMT1 | Glioma | Down | [152][39] |
| miR-185 | DNMT1 | Glioma | Down | [153][40] |
| miR-145 | DNMT3A | Ovarian | Down | [144][41] |
| miR-101 | DNMT3A | Glioma | Down | [154][42] |
| miR-29 | DNMT3B | Burkitt | Down | [142][43] |
| miR-29b | DNMT3B | Lymphoma, pancreatic, head and neck cell line cancer |
Down | [143,155][44][45] |
References
- Ponger, L.; Li, W.-H. Evolutionary Diversification of DNA Methyltransferases in Eukaryotic Genomes. Mol. Biol. Evol. 2005, 22, 1119–1128.
- Campos, C.; Valente, L.M.P.; Fernandes, J.M.O. Molecular Evolution of Zebrafish Dnmt3 Genes and Thermal Plasticity of Their Expression during Embryonic Development. Gene 2012, 500, 93–100.
- Mosquera-Rendón, J.; Cárdenas-Brito, S.; Pineda, J.D.; Corredor, M.; Benítez-Páez, A. Evolutionary and Sequence-Based Relationships in Bacterial AdoMet-Dependent Non-Coding RNA Methyltransferases. BMC Res. Notes 2014, 7, 440.
- Zhenilo, S.V.; Sokolov, A.S.; Prokhortchouk, E.B. Epigenetics of Ancient DNA. Acta Nat. 2016, 30, 72–76.
- Jurkowski, T.P.; Jeltsch, A. On the Evolutionary Origin of Eukaryotic DNA Methyltransferases and Dnmt2. PLoS ONE 2011, 6, e28104.
- Bheemanaik, S.; Reddy, Y.V.R.; Rao, D.N. Structure, Function and Mechanism of Exocyclic DNA Methyltransferases. Biochem. J. 2006, 399, 177–190.
- Chédin, F. The DNMT3 Family of Mammalian De Novo DNA Methyltransferases. In Progress in Molecular Biology and Translational Science; Elsevier: Amsterdam, The Netherlands, 2011; Volume 101, pp. 255–285.
- Jurkowska, R.Z.; Jurkowski, T.P.; Jeltsch, A. Structure and Function of Mammalian DNA Methyltransferases. Chembiochem 2011, 12, 206–222.
- Lee, J.-H.; Voo, K.S.; Skalnik, D.G. Identification and Characterization of the DNA Binding Domain of CpG-Binding Protein. J. Biol. Chem. 2001, 276, 44669–44676.
- Edwards, J.R.; Yarychkivska, O.; Boulard, M.; Bestor, T.H. DNA Methylation and DNA Methyltransferases. Epigenetics Chromatin 2017, 10, 23.
- Xu, T.-H.; Liu, M.; Zhou, X.E.; Liang, G.; Zhao, G.; Xu, H.E.; Melcher, K.; Jones, P.A. Structure of Nucleosome-Bound DNA Methyltransferases DNMT3A and DNMT3B. Nature 2020, 586, 151–155.
- Taverna, S.D.; Li, H.; Ruthenburg, A.J.; Allis, C.D.; Patel, D.J. How Chromatin-Binding Modules Interpret Histone Modifications: Lessons from Professional Pocket Pickers. Nat. Struct. Mol. Biol. 2007, 14, 1025–1040.
- Bestor, T.H. Cytosine Methylation Mediates Sexual Conflict. Trends Genet. 2003, 19, 185–190.
- Goyal, R. Accuracy of DNA Methylation Pattern Preservation by the Dnmt1 Methyltransferase. Nucleic Acids Res. 2006, 34, 1182–1188.
- Fatemi, M.; Hermann, A.; Pradhan, S.; Jeltsch, A. The Activity of the Murine DNA Methyltransferase Dnmt1 Is Controlled by Interaction of the Catalytic Domain with the N-Terminal Part of the Enzyme Leading to an Allosteric Activation of the Enzyme after Binding to Methylated DNA. J. Mol. Biol. 2001, 309, 1189–1199.
- Li, E.; Zhang, Y. DNA Methylation in Mammals. Cold Spring Harb. Perspect. Biol. 2014, 6, a019133.
- Feinberg, A.P.; Vogelstein, B. Hypomethylation Distinguishes Genes of Some Human Cancers from Their Normal Counterparts. Nature 1983, 301, 89–92.
- Feinberg, A.P.; Vogelstein, B. A Technique for Radiolabeling DNA Restriction Endonuclease Fragments to High Specific Activity. Anal. Biochem. 1983, 132, 6–13.
- Ley, T.J.; Ding, L.; Walter, M.J.; McLellan, M.D.; Lamprecht, T.; Larson, D.E.; Kandoth, C.; Payton, J.E.; Baty, J.; Welch, J.; et al. DNMT3A Mutations in Acute Myeloid Leukemia. N. Engl. J. Med. 2010, 363, 2424–2433.
- Kanai, Y.; Ushijima, S.; Nakanishi, Y.; Sakamoto, M.; Hirohashi, S. Mutation of the DNA Methyltransferase (DNMT) 1 Gene in Human Colorectal Cancers. Cancer Lett. 2003, 192, 75–82.
- Amatori, S.; Bagaloni, I.; Donati, B.; Fanelli, M. DNA Demethylating Antineoplastic Strategies: A Comparative Point of View. Genes Cancer 2010, 1, 197–209.
- Huang, D.; Cui, L.; Ahmed, S.; Zainab, F.; Wu, Q.; Wang, X.; Yuan, Z. An Overview of Epigenetic Agents and Natural Nutrition Products Targeting DNA Methyltransferase, Histone Deacetylases and MicroRNAs. Food Chem. Toxicol. 2018, 123, 574–594.
- Pan, Y.; Liu, G.; Zhou, F.; Su, B.; Li, Y. DNA Methylation Profiles in Cancer Diagnosis and Therapeutics. Clin. Exp. Med. 2018, 18, 1–14.
- Gnyszka, A.; Jastrzebski, Z.; Flis, S. DNA Methyltransferase Inhibitors and Their Emerging Role in Epigenetic Therapy of Cancer. Anticancer Res. 2013, 33, 2989–2996.
- Daher-Reyes, G.S.; Merchan, B.M.; Yee, K.W.L. Guadecitabine (SGI-110): An Investigational Drug for the Treatment of Myelodysplastic Syndrome and Acute Myeloid Leukemia. Expert Opin. Investig. Drugs 2019, 28, 835–849.
- Datta, J.; Ghoshal, K.; Denny, W.A.; Gamage, S.A.; Brooke, D.G.; Phiasivongsa, P.; Redkar, S.; Jacob, S.T. A New Class of Quinoline-Based DNA Hypomethylating Agents Reactivates Tumor Suppressor Genes by Blocking DNA Methyltransferase 1 Activity and Inducing Its Degradation. Cancer Res. 2009, 69, 4277–4285.
- Rilova, E.; Erdmann, A.; Gros, C.; Masson, V.; Aussagues, Y.; Poughon-Cassabois, V.; Rajavelu, A.; Jeltsch, A.; Menon, Y.; Novosad, N.; et al. Design, Synthesis and Biological Evaluation of 4-Amino-N-(4-Aminophenyl)Benzamide Analogues of Quinoline-Based SGI-1027 as Inhibitors of DNA Methylation. Chemmedchem 2014, 9, 590–601.
- Zwergel, C.; Schnekenburger, M.; Sarno, F.; Battistelli, C.; Manara, M.C.; Stazi, G.; Mazzone, R.; Fioravanti, R.; Gros, C.; Ausseil, F.; et al. Identification of a Novel Quinoline-Based DNA Demethylating Compound Highly Potent in Cancer Cells. Clin. Epigenetics 2019, 11, 68.
- Hu, C.; Liu, X.; Zeng, Y.; Liu, J.; Wu, F. DNA Methyltransferase Inhibitors Combination Therapy for the Treatment of Solid Tumor: Mechanism and Clinical Application. Clin. Epigenetics 2021, 13, 166.
- Ahuja, N.; Sharma, A.R.; Baylin, S.B. Epigenetic Therapeutics: A New Weapon in the War Against Cancer. Annu. Rev. Med. 2016, 67, 73–89.
- Bhat, S.A.; Ahmad, S.M.; Mumtaz, P.T.; Malik, A.A.; Dar, M.A.; Urwat, U.; Shah, R.A.; Ganai, N.A. Long Non-Coding RNAs: Mechanism of Action and Functional Utility. Non-Coding RNA Res. 2016, 1, 43–50.
- Klisovic, R.B.; Stock, W.; Cataland, S.; Klisovic, M.I.; Liu, S.; Blum, W.; Green, M.; Odenike, O.; Godley, L.; Burgt, J.V.; et al. A Phase I Biological Study of MG98, an Oligodeoxynucleotide Antisense to DNA Methyltransferase 1, in Patients with High-Risk Myelodysplasia and Acute Myeloid Leukemia. Clin. Cancer Res. 2008, 14, 2444–2449.
- Winquist, E.; Knox, J.; Ayoub, J.-P.; Wood, L.; Wainman, N.; Reid, G.K.; Pearce, L.; Shah, A.; Eisenhauer, E. Phase II Trial of DNA Methyltransferase 1 Inhibition with the Antisense Oligonucleotide MG98 in Patients with Metastatic Renal Carcinoma: A National Cancer Institute of Canada Clinical Trials Group Investigational New Drug Study. Investig. New Drugs 2006, 24, 159–167.
- Plummer, R.; Vidal, L.; Griffin, M.; Lesley, M.; de Bono, J.; Coulthard, S.; Sludden, J.; Siu, L.L.; Chen, E.X.; Oza, A.M.; et al. Phase I Study of MG98, an Oligonucleotide Antisense Inhibitor of Human DNA Methyltransferase 1, Given as a 7-Day Infusion in Patients with Advanced Solid Tumors. Clin. Cancer Res. 2009, 15, 3177–3183.
- Qi, D.; Li, J.; Que, B.; Su, J.; Li, M.; Zhang, C.; Yang, M.; Zhou, G.; Ji, W. Long Non-Coding RNA DBCCR1-003 Regulate the Expression of DBCCR1 via DNMT1 in Bladder Cancer. Cancer Cell Int. 2016, 16, 81.
- Li, W.; Zheng, J.; Deng, J.; You, Y.; Wu, H.; Li, N.; Lu, J.; Zhou, Y. Increased Levels of the Long Intergenic Non-Protein Coding RNA POU3F3 Promote DNA Methylation in Esophageal Squamous Cell Carcinoma Cells. Gastroenterology 2014, 146, 1714–1726.
- Yan, J.; Guo, X.; Xia, J.; Shan, T.; Gu, C.; Liang, Z.; Zhao, W.; Jin, S. MiR-148a Regulates MEG3 in Gastric Cancer by Targeting DNA Methyltransferase 1. Med. Oncol. 2014, 31, 879.
- Braconi, C.; Kogure, T.; Valeri, N.; Huang, N.; Nuovo, G.; Costinean, S.; Negrini, M.; Miotto, E.; Croce, C.M.; Patel, T. MicroRNA-29 Can Regulate Expression of the Long Non-Coding RNA Gene MEG3 in Hepatocellular Cancer. Oncogene 2011, 30, 4750–4756.
- Zhang, P.; Sun, H.; Yang, B.; Luo, W.; Liu, Z.; Wang, J.; Zuo, Y. MiR-152 Regulated Glioma Cell Proliferation and Apoptosis via Runx2 Mediated by DNMT1. Biomed. Pharmacother. 2017, 92, 690–695.
- Zhang, Z.; Tang, H.; Wang, Z.; Zhang, B.; Liu, W.; Lu, H.; Xiao, L.; Liu, X.; Wang, R.; Li, X.; et al. MiR-185 Targets the DNA Methyltransferases 1 and Regulates Global DNA Methylation in Human Glioma. Mol. Cancer 2011, 10, 1–16.
- Zhang, S.; Pei, M.; Li, Z.; Li, H.; Liu, Y.; Li, J. Double-negative Feedback Interaction between DNA Methyltransferase 3A and MicroRNA-145 in the Warburg Effect of Ovarian Cancer Cells. Cancer Sci. 2018, 109, 2734–2745.
- Liu, X.; Lei, Q.; Yu, Z.; Xu, G.; Tang, H.; Wang, W.; Wang, Z.; Li, G.; Wu, M. MiR-101 Reverses the Hypomethylation of the LMO3 Promoter in Glioma Cells. Oncotarget 2015, 6, 7930.
- Mazzoccoli, L.; Robaina, M.C.; Apa, A.G.; Bonamino, M.; Pinto, L.W.; Queiroga, E.; Bacchi, C.E.; Klumb, C.E. MiR-29 Silencing Modulates the Expression of Target Genes Related to Proliferation, Apoptosis and Methylation in Burkitt Lymphoma Cells. J. Cancer Res. Clin. Oncol. 2018, 144, 483–497.
- Wang, L.; Huang, J.; Wu, C.; Huang, L.; Cui, J.; Xing, Z.; Zhao, C. Downregulation of MiR-29b Targets DNMT3b to Suppress Cellular Apoptosis and Enhace Proliferation in Pancreatic Cancer. Mol. Med. Rep. 2018, 17, 2113–2120.
- Chen, L.-H.; Hsu, W.-L.; Tseng, Y.-J.; Liu, D.-W.; Weng, C.-F. Involvement of DNMT 3B Promotes Epithelial-Mesenchymal Transition and Gene Expression Profile of Invasive Head and Neck Squamous Cell Carcinomas Cell Lines. BMC Cancer 2016, 16, 431.
- Jones, R.; Wijesinghe, S.; Wilson, C.; Halsall, J.; Liloglou, T.; Kanhere, A. A Long Intergenic Non-Coding RNA Regulates Nuclear Localization of DNA Methyl Transferase-1. Iscience 2021, 24, 102273.
- Somasundaram, S.; Forrest, M.E.; Moinova, H.; Cohen, A.; Varadan, V.; LaFramboise, T.; Markowitz, S.; Khalil, A.M. The DNMT1-Associated LincRNA DACOR1 Reprograms Genome-Wide DNA Methylation in Colon Cancer. Clin. Epigenetics 2018, 10, 127.
- Guo, X.; Chen, Z.; Zhao, L.; Cheng, D.; Song, W.; Zhang, X. Long Non-Coding RNA-HAGLR Suppressed Tumor Growth of Lung Adenocarcinoma through Epigenetically Silencing E2F1. Exp. Cell Res. 2019, 382, 111461.
More
