The gut microbiome, or gut microbiota, also termed commensal, refers to the entire microbial community that populates the mammalian gastrointestinal (GI) tract, with the majority residing in the colon. Alterations of the gut microbiome are implicated in many gastrointestinal diseases, such as inflammatory bowel disease (IBD). Food components in our diet provide not only nutrients to our body but also substrates for the gut microbial flora. What we eat shapes the structure, composition, and function of the gut microbiome, which interacts with the gut epithelium and mucosal immune system and maintains intestinal homeostasis in a healthy state. There is growing interest in nutritional therapy to target the gut microbiome in IBD. This review summarizes the current knowledge regarding the impacts of major food components and their metabolites on the gut microbiome and health consequences within the GI tract. Understanding the influence of foods and nutrition on the gut microbiome-host immune system interactions will be helpful in developing nutritional strategies for maintaing gut health and restore a healthy microbiome in IBD.
- gut microbiome
- gut microbiota
- nutrition
- foods
- dietary fiber
1. Introduction
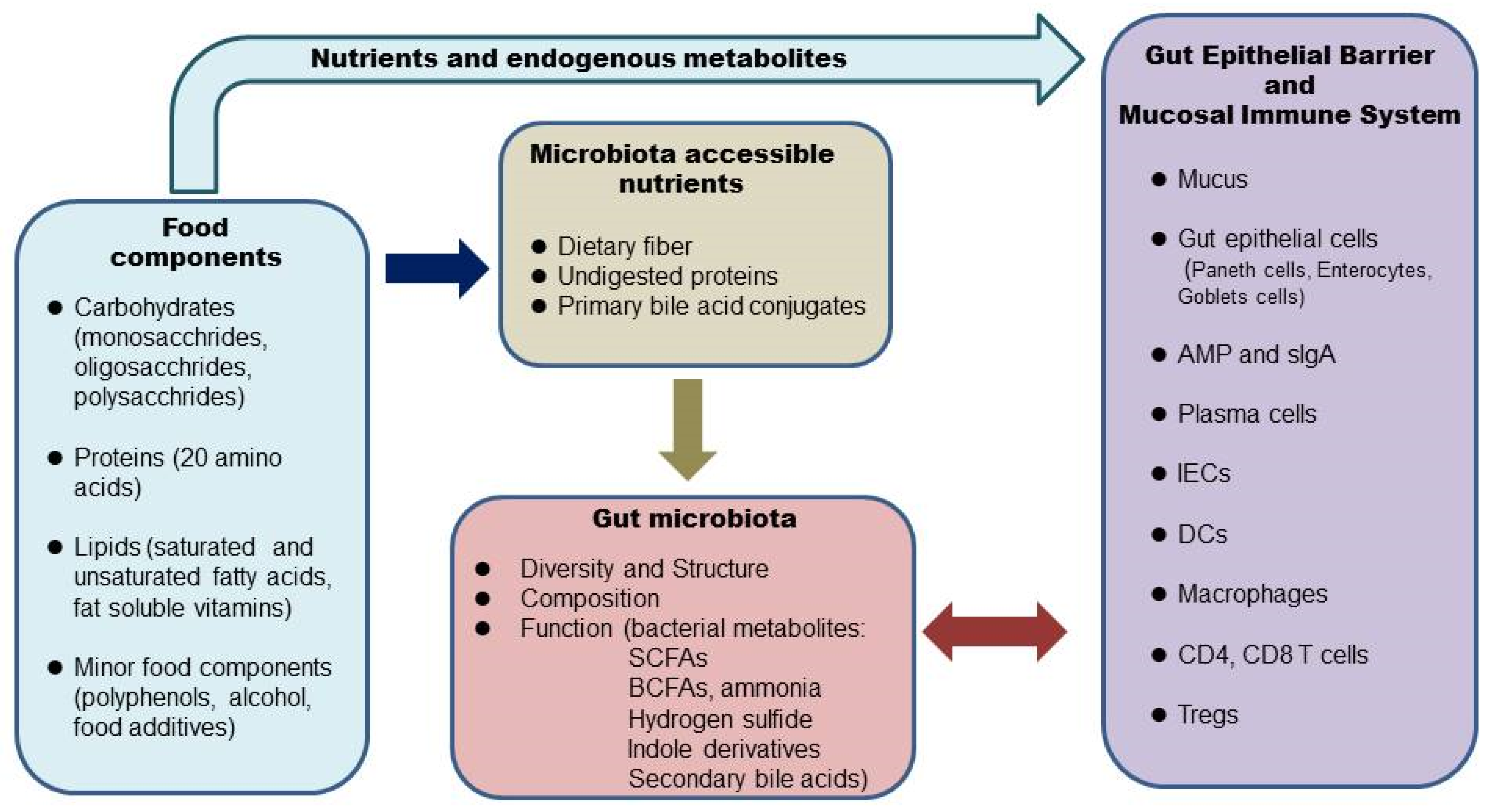
2. Diet, the Microbiome, and the Intestinal Barrier
The human GI tract is covered by a single layer of epithelial cells held together by tight junction proteins such as claudins, occludins, and zonulae occudens (ZO) [15]. The intestinal epithelial cells form a physical barrier as they are impermeable to luminal contents. There are at least seven types of intestinal epithelial cells: enterocytes, goblet cells, Paneth cells, microfold cells, enteroendocrine cells, cup cells, and tuft cells [16]. Enterocytes are the most abundant cells responsible for nutrient uptake [17]. Goblet cells, with more abundance in the distal direction, are responsible for producing mucus [17]. Most Paneth cells reside in the small intestine and secret antimicrobial peptides [17]. The intestinal epithelial cells and the secreted factors form the intestinal barrier [15]. The glycoprotein-rich mucus layer overlying the gut epithelium is the first line of defense against commensal microbes as well as pathogens [17]. MUC2 is the major component of the gel-like mucins in the intestine. The large intestine has two layers of mucus, namely, a firmly attached bacteria-free inner layer and a loose outer layer [17]. The inner layer is about 50 μm thick in mice and 200–300 μm thick in humans. The outer layer expands 4–5 times in volume, which creates a habitat for the commensal bacteria. The mucus barrier is also a reservoir of antimicrobial peptides and IgA. The inner mucus layer is continuously renewed every 1–2 h in murine colonic tissue. Once the inner mucus layer is lost or becomes penetrable to bacteria, a large number of bacteria will reach the epithelial cells and trigger inflammation. Thus, a penetrable inner mucus layer allowing large quantities of bacteria to reach the epithelial cells is a common mechanism for all mouse models of colitis and patients with active ulcerative colitis [17]. Bacterial stimulation is essential for the development and function of the intestinal barrier. In germ-free mice, the mucus layer is extremely thin [18]. The permeability of the intestinal barrier is tightly regulated in a healthy gut. The commensal bacteria maintain the epithelial barrier by providing energy in the form of short-chain fatty acids and also releasing antimicrobial substances to inhibit pathogens. Some nutrients are important regulators of tight junction protein levels, which are critical in maintaining the epithelial barrier [15]. An increase in intestinal permeability, termed a “leaky” gut, can be induced by dietary factors and may trigger inflammatory responses [19]. In a healthy gut, a balance exists between commensal bacteria and the mucus layer. Some gut bacteria, termed mucin specialists, specifically metabolize mucins and are the major mucin degraders when the diet is rich in dietary polysaccharides. There is a balance of production and degradation of mucus, which maintains the thickness of the mucus layer. Dietary fiber-derived SCFAs promote the integrity of intestinal epithelium by inducing goblet cells to increase mucin production [2] and enterocytes to secret IL-18, which is important for epithelial repair [20]. SCFAs can also directly modify tight junctions to strengthen the gut barrier [15]. When the diet is devoid of dietary fibers, some mucin generalists switch metabolism from plant polysaccharides to host mucin glycans. Expansion of mucus-degrading bacteria and an increase in the metabolic activity in utilizing mucin glycans lead to erosion of the mucus layer [18]. Reduced dietary fiber correlates with the thinning of colonic mucus. Different protein sources also affect the thickness of the mucus layer [21]. High saturated fats impair intestinal barrier integrity by reducing tight junction protein occludin and ZO-1 [22,23][22][23]. Simple sugars [24,25][24][25] and emulsifiers [26] negatively affect the intestinal barrier by inducing the expansion of mucin lytic bacteria such as Akkermansia muciniphila, which leads to a thinning of the mucus layer. Some food components (milk fat) promote the growth of Proteobacteria, which produces compounds that are toxic to the intestinal epithelial cells [26]. A leaky gut is involved in the pathogenesis of many inflammatory diseases, including IBD [19].3. Diet, the Microbiome, and the Intestinal Mucosal Immune System
Underneath the intestinal epithelial layer is the lamina propria, where most of the intestinal mucosal immune system resides [17]. Here, various types of innate and adaptive immune cells are found: dendritic cells, macrophages, innate lymphoid cells (ILCs), CD4+ T cells (Th1, Th17, Treg cells), CD8+ T cells, and IgA-secreting plasma cells. These cells work in concert in defense against pathogen infection and in the maintenance of the intestinal mucosal barrier. Unrestrained inflammatory responses to food antigens or commensal bacteria are the main causes of chronic intestinal inflammation and tissue damage in human IBD patients [12]. Under normal conditions, the mucosal immune system is tightly regulated. Local Tregs play a critical role in colon homeostasis [27,28][27][28]. Many bacterial metabolites induce colonic Tregs, such as SCFAs, certain secondary bile acid conjugates, and tryptophan metabolites [29,30,31,32,33][29][30][31][32][33]. The commensal bacteria and the immune system evolve and interplay with each other. Diet influences this interplay by providing substrates for the gut bacteria, and some nutrients can directly modulate immune cells. Normal development and function of the immune system depend on bacterial stimulation. Germ-free mice show defects in several immune cells and are more susceptible to infection [34]. In mice monocolonized with human gut microbes, immune responses show diversity and redundancy [34]. Most microbes elicit distinct and shared responses at both transcriptional and cellular levels. The broad and redundant immune changes induced by gut microbes provide a consistent impact on the host and promote overall health. A recent human study showed that a diet rich in fermented foods leads to increased microbial diversity and decreases in numerous markers of inflammation [10]. The effect is probably through modulations in the gut microbes and metabolites. It is well established that Foxp3+ Treg cells play a central role in the maintenance of immune homeostasis and particularly in the intestine. This is a subset of CD4+CD25+ T cells expressing the transcription factor Forkhead box P3 (Foxp3), which could suppress spontaneous multi-organ autoimmunity, including gastrointestinal inflammation induced by CD4+CD25− T cells [35]. Tregs represent around 10% of CD4+ T cells and were initially discovered to present only in lymphoid tissues; however, recent studies showed the existence of tissue Tregs [36]. Two colonic Treg populations have been identified: one comes from the thymus and proliferates in the colon expressing Helios and Gata3; the other one newly differentiates from naïve Foxp3− CD4+ T cells and becomes Helios−RORγt+ [27]. These colonic Tregs are as effective as lymphatic tissue Tregs in terms of suppression of effector T cells, thus controlling local inflammation. Another distinctive role of colonic Tregs is involved in local mucosal barrier repair. Colonic Tregs have been shown to suppress symptoms in multiple models of colitis [29,30,31,32][29][30][31][32]. The commensal microbes play a major role in shaping the population of Foxp3+CD4+ T regulator cells in the colon. Colonic Tregs are reduced in germ-free mice or following antibiotic treatment. A number of individual gut microbes strongly induce colonic Tregs, including Clostridia clusters IV, XIVa and XVIII, and some Bacteroides species [28]. These bacteria produce SCFAs by fermentation of dietary fiber. The very low number of colonic Tregs in germ-free mice can be rescued by acetate, propionate, or butyrate, indicating these SCFAs work independently [24]. Different SCFAs induce colonic Treg population through multiple mechanisms. For example, acetate promotes the expansion of pre-existing colonic Tregs by activation of FFAR2 on T cells, whereas butyrate increases the de novo differentiation of colonic Tregs by inhibiting histone deacetylase (HDAC) activity [30]. SCFAs also indirectly promote colonic Tregs expansion by affecting DC maturation through activation of GPR109A on DCs [29].References
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533.
- Postler, T.S.; Ghosh, S. Understanding the holobiont: How microbial metabolites affect human health and shape the immune system. Cell Metab. 2017, 26, 110–130.
- Arumugam, M.; Raes, J.; Pelletier, E.; le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180.
- Frank, D.N.; St. Amand, A.L.; Feidman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785.
- Schroeder, B.; Bäckhed, F. Signals from the gut microbiota to distant organs in physiology and disease. Nat. Med. 2016, 22, 1079–1089.
- Singh, R.K.; Chang, H.W.; Yan, D.; Lee, K.M.; Ucmak, D.; Wong, K.; Abrouk, M.; Farahnik, B.; Nakamura, M.; Zhu, T.H.; et al. Influence of diet on the gut microbiome and implications for human health. J. Transl. Med. 2017, 15, 73.
- Smits, S.A.; Leach, J.; Sonnenburg, E.D.; Gonzalez, C.G.; Lichtman, J.S.; Reid, G.; Knight, R.; Manjurano, A.; Changalucha, J.; Elias, J.E.; et al. Seasonal cycling in the gut microbiome of the Hadaz hunter-gatherers of Tanzania. Science 2016, 357, 802–806.
- Jha, A.R.; Davenport, E.R.; Gautam, Y.; Bhandari, D.; Tandukar, S.; Ng, K.M.; Fragiadakis, G.K.; Holmes, S.; Gautam, G.P.; Leach, J.; et al. Gut microbiome transition across a lifestyle gradient in Himalaya. PLoS Biol. 2018, 16, e2005396.
- Deehan, E.C.; Yang, C.; Perez-Muñoz, M.E.; Nguyen, N.K.; Cheng, C.C.; Triador, L.; Zhang, Z.; Bakal, J.A.; Walter, J. Precision microbiome modulation with discrete dietary fiber structures directs short-chain fatty acids production. Cell Host Microbe 2020, 27, 389–404.
- Wastyk, H.; Fragiadakis, G.K.; Perelman, D.; Dahan, D.; Merrill, B.D.; Yu, F.B.; Topf, M.; Gonzalez, C.G.; Treuren, W.V.; Han, S. Gut-microbiota-targeted diets modulate human immune status. Cell 2021, 184, 4137–4153.
- Rinninella, E.; Cintoni, M.; Raoul, P.; Lopetuso, L.R.; Scaldafferri, F.; Pulcini, G.; Miggiano, G.A.D.; Gasbarrini, A.; Mele, G.P. Food components and dietary habits: Keys for a healthy gut microbiota composition. Nutrients 2019, 11, 2393.
- Lane, E.R.; Zisman, T.L.; Suskind, D.L. The microbiota in inflammatory bowel disease: Current and therapeutic insights. J. Inflamm. Res. 2017, 10, 63–73.
- Sugihara, K.; Kamada, N. Diet-microbiota interactions in inflammatory bowel disease. Nutrients 2021, 13, 1533.
- Staley, C.; Weingarden, A.R.; Khoruts, A.; Sadowsky, M.J. Interaction of gut microbiota with bile acid metabolism and its influence on disease states. Appl. Microbiol. Biotechnol. 2017, 101, 47–64.
- De Santis, S.; Cavalcanti, E.; Mastronardi, M.; Jirillo, E.; Chieppa, M. Nutritional keys for intestinal barrier modulation. Front. Immunol. 2015, 6, 612.
- Gerbe, F.; Legraverend, C.; Jay, P. The intestinal epithelium tuft cells: Specification and function. Cell Mol. Life Sci. 2012, 69, 2907–2917.
- Pelaseyed, T.; Bergström, J.H.; Gustafsson, J.K.; Ermund, A.; Birchenough, G.M.H.; Schütte, A.; van der Post, S.; Svensson, F.; Rodríguez-Piñeiro, A.M.; Nyström, E.E.L.; et al. The mucus and mucins of the goblet cells and enterocytes provide the first defense line of the gastrointestinal tract and interact with the immune system. Immonol. Rev. 2014, 260, 8–20.
- Desal, M.S.; Seekatz, A.M.; Koropatkin, N.K.; Kamada, N.; Hickey, C.A.; Wolter, M.; Pudlo, N.A.; Kitamoto, S.; Terrapon, N.; Muller, A.; et al. A dietary fiber-deprived gut microbiota degrades the colonic mucus barrier and enhances pathogen susceptibility. Cell 2016, 167, 1339–1353.
- Mu, Q.; Kirby, J.; Reilly, C.M.; Luo, X.M. Leaky gut as a danger signal for autoimmune diseases. Front. Immonol. 2017, 8, 598.
- Kalina, U.; Koyama, N.; Hosoda, T.; Nuernberger, H.; Sato, K.; Hoelzer, D.; Herweck, F.; Manigold, T.; Singer, M.V.; Rossol, S.; et al. Enhanced production of IL-18 in butyrate-treated intestinal epithelium by stimulation of the proximal promoter region. Eur. J. Immunol. 2002, 32, 2635–2643.
- Zhao, F.; Zhou, G.; Liu, X.; Song, S.; Xu, X.; Hooiveld, G.; Müller, M.; Liu, L.; Kristiansen, K.; Li, C. Dietary protein sources differentially affect the growth of akkermansia muciniphila and maintenance of the gut mucus barrier in mice. Mol. Nutr. Food Res. 2019, 63, 1900589.
- Gruber, L.; Kisling, S.; Lichti, P.; Martin, F.P.; May, S.; Klingenspor, M.; Lichtenegger, M.; Rychlik, M.; Haller, D. High fat diet accelerates pathogenesis of murine crohn’s disease-like ileitis independently of obesity. PLoS ONE 2013, 8, e71161.
- Lam, Y.Y.; Ha, C.W.Y.; Campbell, C.R.; Mitchell, A.J.; Dinudom, A.; Oscarsson, J.; Cook, D.I.; Hunt, N.H.; Caterson, I.D.; Holmes, A.J.; et al. Increased gut permeability and microbioa change associate with mesenteric fat inflammation and metabolic dysfunction in diet-induced obese mice. PLoS ONE 2012, 7, e34233.
- Khan, S.; Waliullah, S.; Godfrey, V.; Khan, M.A.W.; Ramachandran, R.A.; Cantarel, B.; Behrendt, C.; Peng, L.; Hooper, L.V.; Zaki, H. Dietary simple sugars alter microbial ecology in the gut and promote colitis in mice. Sci. Transl. Med. 2020, 12, eaay6218.
- Do, M.H.; Lee, E.; Oh, M.J.; Kim, Y.; Park, H.Y. High-glucose or -fructose diet cause changes of the gut microbiota and metabolic disorders in mice without body weight changes. Nutrients 2018, 10, 761.
- Chassaing, B.; Koren, O.; Goodrich, J.K.; Poole, A.C.; Srinivasan, S.; Ley, R.E.; Gewirtz, A.T. Dietary emulsifiers impact the mouse gut microbiota promoting colitis and metabolic syndrome. Nature 2015, 519, 92–96.
- Panduro, M.; Benoist, C.; Mathis, D. Tissue-Tregs. Annu. Rev. Immunol. 2016, 34, 609–633.
- Tanoue, T.; Atarashi, K.; Honda, K. Development and maintenance of intestinal regulatory T cells. Nat. Rev. Immunol. 2016, 16, 295–309.
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The microbial metabolites, short-chain fatty acids, regulate colonic Treg cell homeostasis. Science 2013, 341, 569–573.
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446–450.
- Campbell, C.; McKenney, P.T.; Konstantinovsky, D.; Isaeva, O.I.; Schizas, M.; Verter, J.; Mai, C.; Jin, W.B.; Guo, C.J.; Violante, S. Bacterial metabolism of bile acids promotes generation of peripheral regulatory T cells. Nature 2022, 581, 475–479.
- Song, X.; Sun, X.; Oh, S.F.; Wu, M.; Zhang, Y.; Zheng, W.; Geva-Zatorysky, N.; Jupp, R.; Mathis, D.; Benoist, C.; et al. Microbial bile acid metabolites modulate gut RORγ+ regulatory T cell homeostasis. Nature 2020, 577, 410–415.
- Lamas, B.; Richard, M.L.; Leducaq, V.; Pham, H.-P.; Michel, M.-L.; da Costa, G.; Bridonneau, C.; Jegou, S.; Hoffmann, T.W.; Natividad, J.M.; et al. CARD9 impacts colitis by altering gut microbiota metabolism of tryptophan into aryl hydrocarbon receptor ligands. Nat. Med. 2016, 22, 598–605.
- Geva-Zatorsky, N.; Sefik, E.; Kua, L.; Pasman, L.; Tan, T.G.; Ortiz-Lopez, A.; Yanortsang, T.B.; Yang, L.; Jupp, R.; Mathis, D. Mining the human gut microbiota for immunomodulatory organisms. Cell 2017, 168, 928–943.
- Fontenot, J.D.; Gavin, M.A.; Rudensky, A.Y. Foxp3 programs the development and function of CD4+CD25+ regulatory T cells. Nat. Immunol. 2003, 4, 330–336.
- Sakai, R.; Komai, K.; Iizuka-Koga, M.; Yoshimura, A.; Ito, M. Regulatory T cells: Pathophysiological roles and clinical applications. Keio J. Med. 2020, 69, 1–15.
