Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by di costanzo Luigi.
Biominerals are extraordinary materials that provide organisms with a variety of functions to support life. The synthesis of biominerals and organization at the macroscopic level is a consequence of the interactions of these materials with proteins. The association of biominerals and proteins is very ancient and has sparked a wealth of research across biological, medical and material sciences. Calcium carbonate, hydroxyapatite, and silica represent widespread natural biominerals. The atomic details of the interface between macromolecules and these biominerals is very intriguing from a chemical perspective, considering the association of chemical entities that are structurally different.
- biominerals
- atomic details
- interactions
- calcium carbonate
- structural biology
- protein design
1. Introduction
Biominerals are extraordinary materials with a variety of functions, including providing rigid support for soft tissue, breaking down food, protection from prey, sheltering, and more. Therefore, biominerals represent an attractive natural source of inspiration for novel bioengineered materials with properties adapted to vital functions [1,2,3,4][1][2][3][4]. From eggshells to pearls, bivalve mollusks to sponges, biominerals offer a variety of materials in nature, and like silica, often is produced on a scale than industrial amount [5,6,7][5][6][7]. Even more astonishing is the fact that these materials are made by organisms in very mild conditions.
Notoriously, chicken eggshells are made of more than >90 w/w% of a calcite mineral (calcium carbonate, CaCO3) in association with ~0.03 w/w% uronic acid (carbohydrate) and small amounts of other amino acids [8]. These associations of biomineral and organic molecules/macromolecules make eggs resistant, but still fragile to allow etching [9,10,11][9][10][11]. Bones contain hydroxyapatite, consisting of a phosphate cluster anion counterbalanced by calcium and the addition of two hydroxy ions (Ca10(PO4)6(OH)2). This biomineral, together with collagen, the most abundant protein in our body, helps to confer our skeleton with strength and elasticity [12].
Organisms are equipped with biominerals, often interspersed with proteins (or other organic molecules), that help to glue tiny inorganic crystals to form biomineral aggregates. From a chemical perspective, these crystals are formed by salts of carbonate, citrate, iron oxide, oxalate, phosphate, silicate, sulfates, etc., often counterbalanced by cations of calcium, iron, magnesium, sodium, etc. Biomineral crystals in turn aggregate and assemble to form defined functional shapes ranging from beautiful spherical pearls and amazing spirals of snail shelters. Biominerals can even form tiny compasses assembled in strings that confer bacteria (and other organisms) sensing of the directional Earth’s magnetic field and to drive their motion towards nutrients and/or long-distance migration [9,13,14][9][13][14]. Notably, bacteria known as magnetotactic bacteria, a class of gram-negative bacteria, are able to biomineralize iron into magnetic particles of magnetite, a mixed oxo-iron compound (Fe(II)Fe(III)2O4), or greigite, the equivalent biomineral with sulphur atoms (Fe(II)Fe(III)2S4) [13,15,16][13][15][16]. Magnetite aggregates are grown in an enclosed lipid-bilayer membrane organelle known as magnetosomes [15,17,18,19,20,21][15][17][18][19][20][21]. In addition to the key role of crystal nucleation and mineral size regulation, proteins also confer properties such as elasticity and resilience to pressure.
Besides supporting mineral formation, proteins also provide other necessary tools (such as cell organelle membrane formation, etc.) or the cargo transport of inorganic ions for crystal formation. Proteins help to build cellular compartments to host the growing minerals or provide signalling factors for crystal growth. Proteins or designed peptides can stabilize an unusual crystal form aggregation that would otherwise be unstable and help to prepare mineralized mimicking bone structures [22,23][22][23]. Biomineral properties are not only derived from coexistence of different chemical entities, but also the synergy of several biophysical factors (e.g., pH, temperature, concentration, inhibitors concentration) can affect biochemical pathways synthesizing biominerals and can provide further insights into projects for artificial synthesis of biominerals [4,24,25,26][4][24][25][26]. From a chemical perspective, it is very intriguing to study the interactions of chemical entities that are so structurally different.
Considering the growing interest for biominerals and their potential applications as materials obtained from sustainable processes, biomineralization proteins involving all biochemical pathways of biomineral biosynthesis has grown [27]. Structural properties of biological/organic macromolecules affect biomineral properties on a macroscale level and through a common principle based on non-classical crystal growth [28,29][28][29]. In fact, distantly related organisms adapting to their environment generally use a common universal motif (aka given stretch of protein sequence) that in turn is used as a tool to assemble a material on to the macro scale level with shape and size as needed. In other words, the complex variety of shapes of biominerals on the macro scale seems to have a different and less organized level of complexity on the micro scale level [30,31][30][31].
Although determination of the atomic details of proteins in complex with a biomineral is not straightforward, because of the different chemical entities involved, crystallography, NMR, and cryo-electron microscopy have been instrumental in understanding the nature of interactions of biomineralization proteins and biominerals. Despite the growing number of ordered biomineral protein structures in the Protein Data Bank (PDB), the archive of the macromolecular information, the structural biology of these well-ordered macromolecules represents a segment of larger protein sequence space [31]. In fact, a key role is played by biomineralization proteins that are intrinsically disordered (IDP proteins) or contain a sequence region that only becomes structurally ordered/disordered or are able to undergo variable post-translational modification in the presence of inorganic materials [32]. IDP proteins are more difficult to study by conventional structural biology techniques. Among IDP proteins, osteopontin, a protein expressed in our bones and tissues, is highly negatively charged and has multiple functions including bone structuring [33]. Even more insightful, is the role of biomineralization proteins with multifunctional capabilities such as deposit, nucleation, crystal formation and inhibition [34,35,36][34][35][36].
IResearcher have mined the RCSB PDB, a part of the wwPDB, to provide an overall view of the key atomic details of structurally ordered biomineralization proteins, an analysis of the type of nature of interactions between these macromolecules and biominerals [31]. Notably, a number of biomedical and other applications involving biomineralization proteins are rapidly growing [37,38,39][37][38][39]. Therefore, IResearcher have extended outheir search to find the 3D structural details of designed proteins or other known natural proteins, including enzymes, repurposed for the synthesis of novel biominerals. A list of unique experimentally determined protein structures including functions, associated biominerals, bound ligands, and other key features (functional residues) is reported in Table 1.
Table 1. Natural, designed, and re-engineered proteins for the synthesis of functional biominerals. The unique entries are retrieved from the RCSB PDB (RCSB.org) [31,40][31][40]. The 3D structure for each indicated entry and other protein annotations can be explored by searching from the PDB code, indicated in the second column, and browsing the link below. Related entries can be retrieved from the structure summary page of each entry indicated in the column “PDB code ID”. The ligand name column refers to ligand of interest, if bound to the structure, and not any other solvent. Link: https://www.rcsb.org, accessed on 4 July 2022. The MamE structural model is available from the AlphaFold project [41].
| Source | PDB Code ID | Macromolecule | Crystal Material | Ligand of Interest | Function/Fold Type/Active Site Residues |
|---|---|---|---|---|---|
| Porcine | 1q8h [42] | Osteocalcin | Hydroxyapatite | Calcium ion | Modified Glu residue at position 17, 21, and 24. A -S-S- linkage between two protein chains is present |
| Bovine | 1q3m [43] | Osteocalcin | Hydroxyapatite | -- | Dynamic binding study of calcium ions |
| Bovine | 4mzz [44] | Osteocalcin (3 Glu form) | Hydroxyapatite | -- | Regulation of glucose metabolism. Helical propensity and Glu residues at positions 17, 21, and 24 are also of a focus |
| Argyrosomus regius/fish | 1vzm [45] | Osteocalcin | Hydroxyapatite | Magnesium ion | Modified Glu residue at position 17, 21, 24, and 25 |
| Struthio camelus | 4uww [46] | Struthiocalcin-1 | Calcite | -- | α/β fold, C-type lectin. Intramineral protein. Glu63, Glu64, Glu65, Glu66 and Asp67, Asp93, Asp94, Asp95 and Asp96 |
| Gallus gallus | 1gz2 [47] | Ovocleidin-17 | Calcite | -- | α/β fold, C-type lectin. Intramineral protein. Arg103, Lys106, Arg108, Arg109 and Arg117 |
| Flounder | 1wfa [48] | Antifreeze protein (AFP) | Ice | Water molecules | Inhibition growth of ice crystals. A simple alpha-helix |
| Brachyosis rostratus | 2zib [49] | Antifreeze protein (AFP) | Ice | Water molecules | Inhibition growth of ice crystals. It is a C-type lectin protein |
| Pteria Penguin | 5yrf [50] | PPL3-A | Calcite/Pearl | Trehalose | Carbohydrates as mediators for biomineral recognition. Few -S-S- linkages are present. Asp32 and Glu86, interact with calcium ions, and positively charged residues, Lys83, Lys107, Lys118, Arg119, and Arg147 binding to carbonates anion. Aromatic residues are present |
| Sea urchin | 2jyp [35] | Aragonite protein AP7, C-terminal domain (36 residues) | Mollusk shell, nacre formation | -- | Protein-protein interaction and other functions. Contains a Zn binding −Cys−(X)4−Cys− motif. Depending on biomineral type, binding function can be variable |
| Marine sponge Tethya aurantium | 6zq3 [51] | α-silicatein | Silica/sponges | -- | Silic acid condensation. Hydrolase. Functional residues S26, H165, Q20, N185. Silic acid is added to the crescent silica polymer |
| Human (chimeric construct) | 2vhs [52] | cathepsin L | Silica/sponges | Sulphate | Silic acid condensation. Hydrolase. Catalytic mechanism is described [52]. |
| Magnetospirillum gryphiswaldense | 3asf [53] | MamA, TPR like | Magnetite | -- | Forms a homo-oligomeric scaffold for magnetosome associated proteins guidance |
| Magnetospira | 5ho1 [54] | MamB | Magnetite | Mg2+ | Transport activity for protons and iron ions |
| Magnetosome protein | AF-Q2W8Q8-F1 [55] | MamE, HtrA protein | Magnetite | -- | MamE functions as protease, and together with MamD and MamO, regulates crystal formation and size |
| Magnetospirillum gryphiswaldense | 3w5x [56] | MamM | Magnetite | -- | Transport activity for protons and iron ions |
| Magnetospirillum magneticum | 5jyg [57] | Mamk filament, Actin-like atpase | Magnetite | ADP cofactor, Mg2+ | Mamk aligns magnetosomes. It functions as an actin homolog |
| Magnetospirillum magneticum | 5hm9 [58] | MamO | Magnetite | -- | MamO supports metal transport inside the subcellular compartment. Presence of a surface di-His cluster (H148, H263). Promotes crystal nucleation. |
| Magnetospirillum magnetococcus | 4jj0 [59] | MamP | Magnetite | Heme C | Iron(II) oxidation and mineralization. It performs a control ratio Fe(II)/Fe(III) |
| Designed protein | 5chb [60] | nvPizza2-S16H58 | Nano crystal composed of 7 cadmium ions and 12 chloride ions | Tiny crystal composed of 7 cadmium ions and 12 chloride ions | Protein induce assembly of a small nanocrystal |
| Stenotrophomonas maltophilia | 6k1n [61] | γ-lyase smCSE | CdS, ZnS, Ag2S quantum dots | PLP cofactor | Function: lyase. Tyr 110 interacts with the aromatic group of the PLP cofactor contributing to its orientation and enzyme catalysis |
| Escherichia coli | 7mq6 [62] | Maltose binding protein (MBP) | Gold nanoparticle synthesis | Gold(I); Maltose | Repurposed enzyme. Peptide fused within protein sequence for binding of gold (functional residue Met 322). |
| Chicken | 3p64 [63] | Lysozyme C | Gold nanoparticle | 9 gold ions Au(I), Au(III) | Hydrolase. Protein crystals form several gold ions aggregates |
| Reengineered human ferritin | 3es3 [64] | Ferritin heavy chain | Gold nanoparticle | Gold(I) ions | A total of96 non-native cysteines added to the interior of the shell |
| Pyrococcus furiosus | 2x17 [65] | Ferritin homolog | Silver nanoparticle | Silver(I) ions | Natural protein repurposed for silver nanoparticle growth under reduction condition |
| Frog ferritin | 3ka3 [66] | Ferritin | Iron(II)/(III) biomineralization | Mg2+; Cl− | “bucket brigade” pathway for the iron(II) movement towards the catalytic di-iron center |
2. Classical Protein Templates for the Nucleation and Growth of Biominerals
Osteocalcin, one of the most studied biomineralization proteins, provides a reference point for the key interactions of biomineral recognition, and its atomic details have been available since 2003. After collagen, oteocalcin is the second most abundant protein in our body/bones and acts as a hormone to promote bone growth [67]. In bones, it binds to the surface of hydroxyapatite crystals, a mineral composed of calcium and phosphate. The human osteocalcin is a small protein composed of only 49 amino acids with the presence in its primary structure of a modified amino acid, γ-carboxyglutamic acid residue (Gla, a glutamic residue with an extra carboxylic group), at positions 17, 21, and 24. Furthermore, a disulfide bond between two cysteine residues at positions 23 and 29 helps to stabilize the structure. The atomic details of the crystal structure of porcine osteocalcin, shown in Figure 1, reveals how this protein recognizes the inorganic mineral surface: an alpha-helix presents a set of negatively charged amino acids, spaced in a regular pattern with the ability to recognize calcium ions in the crystal mineral that are in turn structurally organized. The presence of doubly charged Gla residues further supports the importance of properly oriented side chains to coordinate calcium ions at the proper distance and geometry and plays a role in the helical propensity of this protein sequence and regulates its affinity for hydroxyapatite [44]. In fact, several calcium ions were seen in the crystal structure, showing a perfectly-matched spacing of amino acids to ions (Figure 1 showing PDB entry 1q8h) [42]. A more recent study based on mass spectrometry has revealed the presence of osteocalcin fragments presenting a fourth γ-carboxylation at residue 25 not previously detected in the structure of the porcine osteocalcin. A similar result emerged from a structural study from a fish osteocalcin [45,68][45][68]. In summary, from the wealth of the structural studies Iresearcher have learned how calcium-hydroxyapatite is recognized by the protein and how the helical propensity/stability also play a role in mineral recognition.
Calcite, a calcium carbonate biomineral, is also synthesized by a protein similar to osteocalcin: struthiocalcin. Eggshells contain calcite, a mineral form of calcium carbonate, with a matrix of protein sandwiched in between. Naturally, CaCO3 exists as different crystal polymorphs (i.e., calcite, aragonite, vaterite). Like osteocalcin, eggshell protein struthiocalcin, shown in Figure 1, (PDB entry 4uww) binds to the surface of the mineral crystals using an array of acidic amino acids that bind calcium, helping to direct the crystal growth of ostrich eggshell formation [46]. Struthiocalcin has a folding shape termed C-type lectin. Similarly, ovocleidin-17, the intramineral protein found in chicken eggs, offers a similar view of electrostatically charged surface protein residues interacting with inorganic material but with a subtle difference. In place of negatively charged acidic residues (glutamic and aspartic residues), it presents a stretch of positively charged residues (arginine and lysine resides, Table 1) [47,69][47][69].
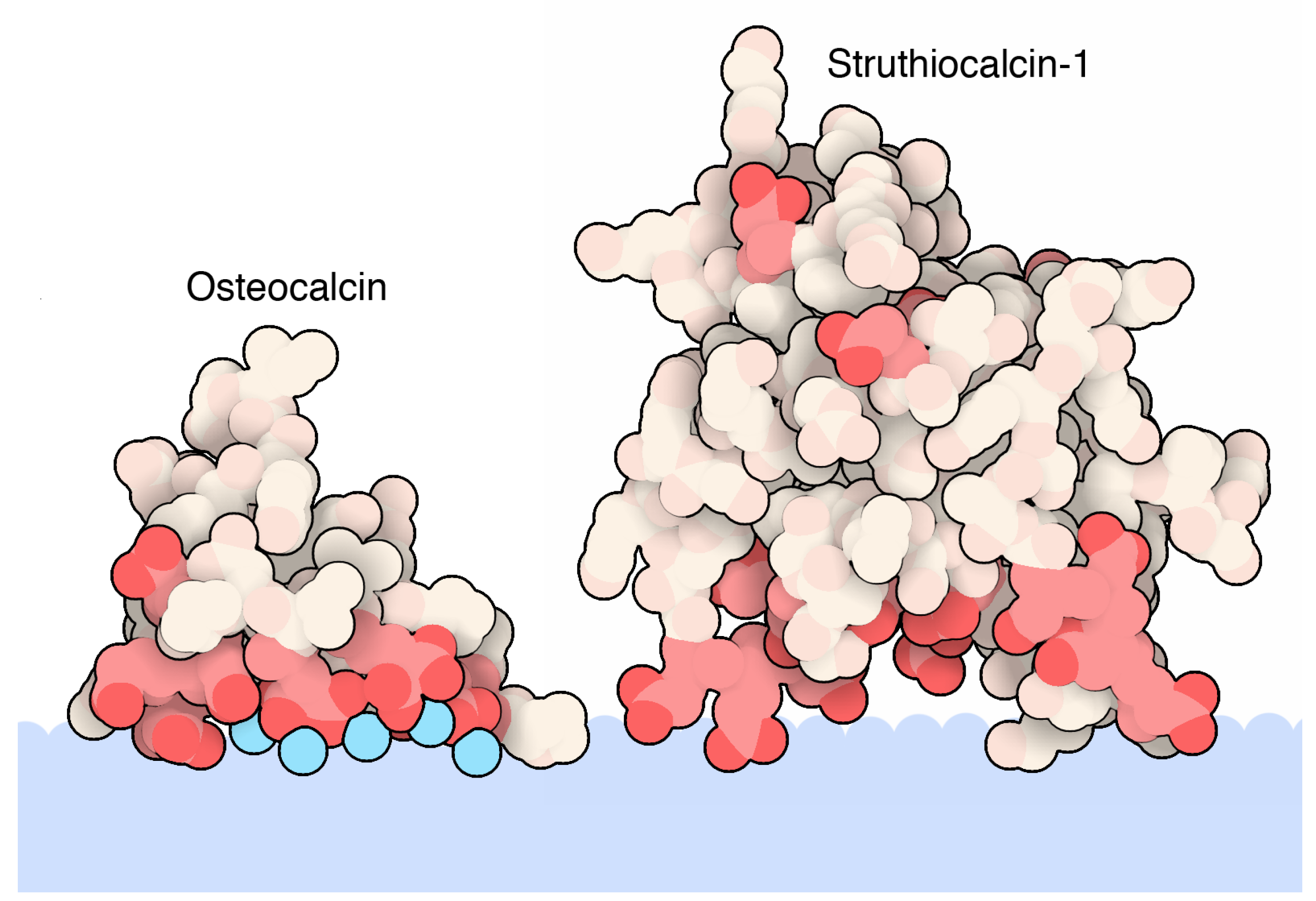
Figure 1. Atomic details of biomineralization proteins binding a calcite biomineral. The biomineal represented as a light blue color shape is interfaced with the rich acidic segment of osteocalcin (PDB accession code 1q8h [42]) and struthiocalcin-1 (PDB accession code 4uww [46]). Both proteins are represented as spheres. Red colors indicate acidic residues (Glu, Gla, and Asp residues). Calcium ions are indicated in cyan colored spheres. This figure is obtained from the Molecule of the Month column “Proteins and Biominerals” (Di Costanzo, L; Goodsell, D. 10.2210/rcsb_pdb/mom_2019_4) [70].
Besides the interest in understanding how calcite material is deposited in eggs, these proteins are also studied for material applications and goose eggshell ansocalcin, an ovocleidin-17 protein, is capable of inducing crystallization of calcite crystals in vitro [71,72][71][72]. Studies of ostrich egg fossils show that the interaction between the mineral and the protein is so strong that fragments of the protein-mineral complex can last for millions of years [73,74][73][74]. Struthiocalcin is a C-type lectin protein, and though it binds to a mineral entity, the shape is similar to antifreeze proteins that bind ice crystals to their surface (Figure 2, PDB entry 2zib), and it is thought to have evolved from a common ancestor [49,75][49][75].
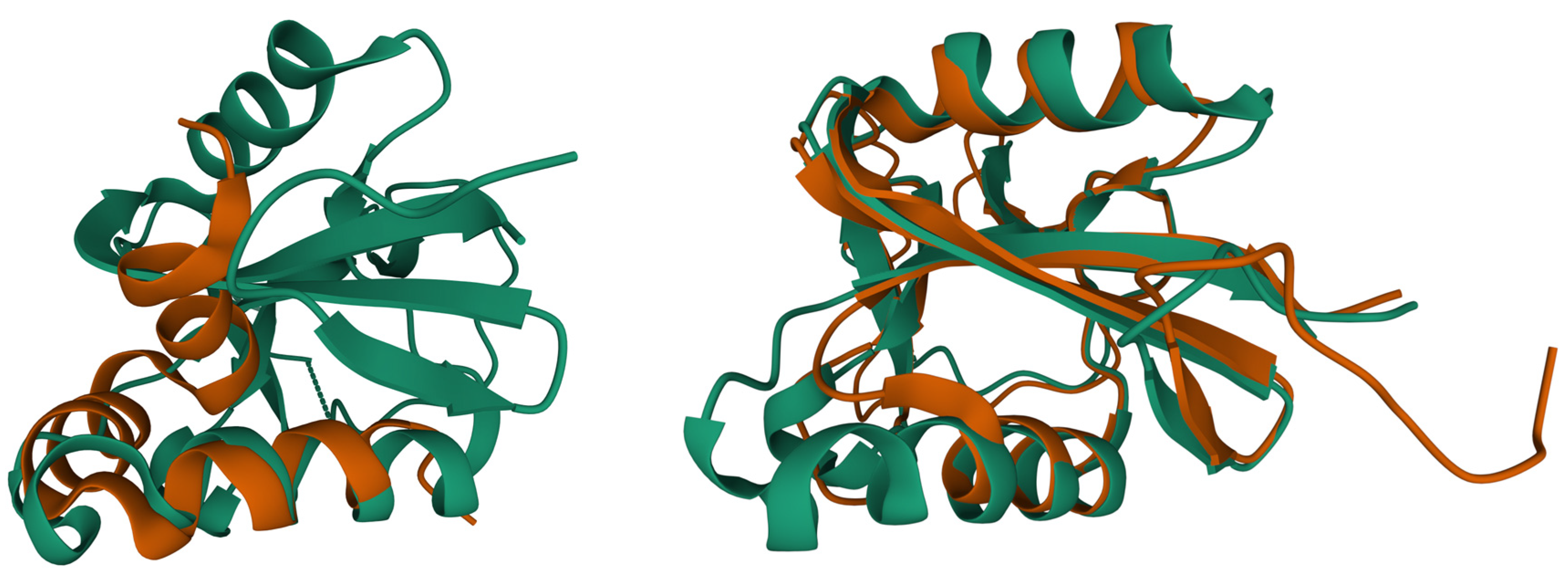
Figure 2. Superposition of biomineralization proteins reveals similarities with the ice binding protein (PDB entry 2zib [49], green color). Superposition of osteocalcin (brown, PDB entry 1q8h [42]) and ice binding protein (left side, rmsd 2.3 Å, 21 Cα atoms). Superposition of struthiocalcin (brown, PDB entry 4uww [46]) and ice binding protein (right side, rmsd 1.5 Å, 119 Cα atoms). Protein structures are represented as ribbon plot. Superpositions were calculated using the Pairwise Structure Alignment tool of the RCSB PDB server.
Even though more than 95% of bivalve shells are composed of calcium carbonate, a quantity less than 5% of an organic matrix reinforces shells mechanical fracture 3000 times as compared to monolithic CaCO3 by allowing the formation of layers with a distinctive pattern to form complex biomineral microstructures [37]. From a biosynthetic perspective, the proteome of oysters’ shell proteins offers insights on how these resilient intertidal zone organisms need to adapt and quickly (even short time) build shells that withstand environmental and climate changes [76]. During the early stage of larva development, typical protein expression includes fibronectin-like protein and chitin synthase, events that are associated with template formation on which crystals of calcium carbonate can form [76]. Generally, the small amount of the organic matrix does influence the mechanical properties of the biomineral and has a key role in controlling the mineralization process, size, crystal morphology, specific crystallographic orientation, polymorph or amorphous phase stabilization, and/or crystal growth inhibition. The studies of protein templates and calcium carbonate is shedding new light on bioceramics and other engineered applications [77,78][77][78].
2.1. Pearls
Among known 3D structures of biomineralization, proteins templating for biomineral growth of the mother pearl formation are of great interest not only to understand how these beautiful structures, which are used for jewelry, are made. Similar to eggshells, oysters’ pearls are made with ~95% calcium carbonate and ~5% of an organic/biological macromolecule such as chitosan and other proteins. Beyond jewelry, mother pearls’ powders are studied for a variety of applications, particularly with regard to health care products, considering their relatively low or no cytotoxicity and osteogenesis stimulation [79].
Upon a stimulus, oysters produce a substance, within a small pearl bag, known as nacre that is made of crystalline layers of calcium carbonate and other organics and biomolecules. The final product is the well-recognized mother pearl. Ongoing research is endeavoring to clarify the mechanism of pearl formation beginning with the nucleation and crystallization of calcium carbonate. This mineral growing into the pearl gem is supported by lectin proteins, a class of biomolecules able to bind sugars. Unlike eggshells, pearl’s calcium carbonate crystal growth is mediated by the binding of sugars, and these in turn could either inhibit or support crystal formation. In vitro crystallization experiments performed with the isoforms of lectin known as PPLs (PPL3A-C and PPL4) have shown a significant effect on the number of calcium carbonate crystals and their size in the presence of sugars. Smaller crystals were increased in a dependent manner of sugar concentration, indicating that its binding might suppress and control crystal growth of calcite [50]. From a structural perspective, as shown in Figure 3, PPL3B 3D-structure to calcite crystals clearly suggest that its interaction on the calcite crystal surface is comprised of negatively charged residues, Asp32 and Glu86, interacting with calcium ions, and positively charged residues, Lys83, Lys107, Lys118, Arg119, and Arg147, binding to carbonates on the crystal faces [5]. Similar conclusions were drawn from the structural analysis of the other PPL3s. The morphology, size, and number of crystals that formed PPL4 are regulated by sugars, resulting in considerably different biomineralization functions, which can either suppress or enhance the crystal growth. Therefore, PPL3 and PPL4 supports calcium carbonate formation or inhibition of mineral formation through carbohydrate binding. Therefore, the mineral recognition mechanism is different with respect to the osteocalcin mechanism (see section above).
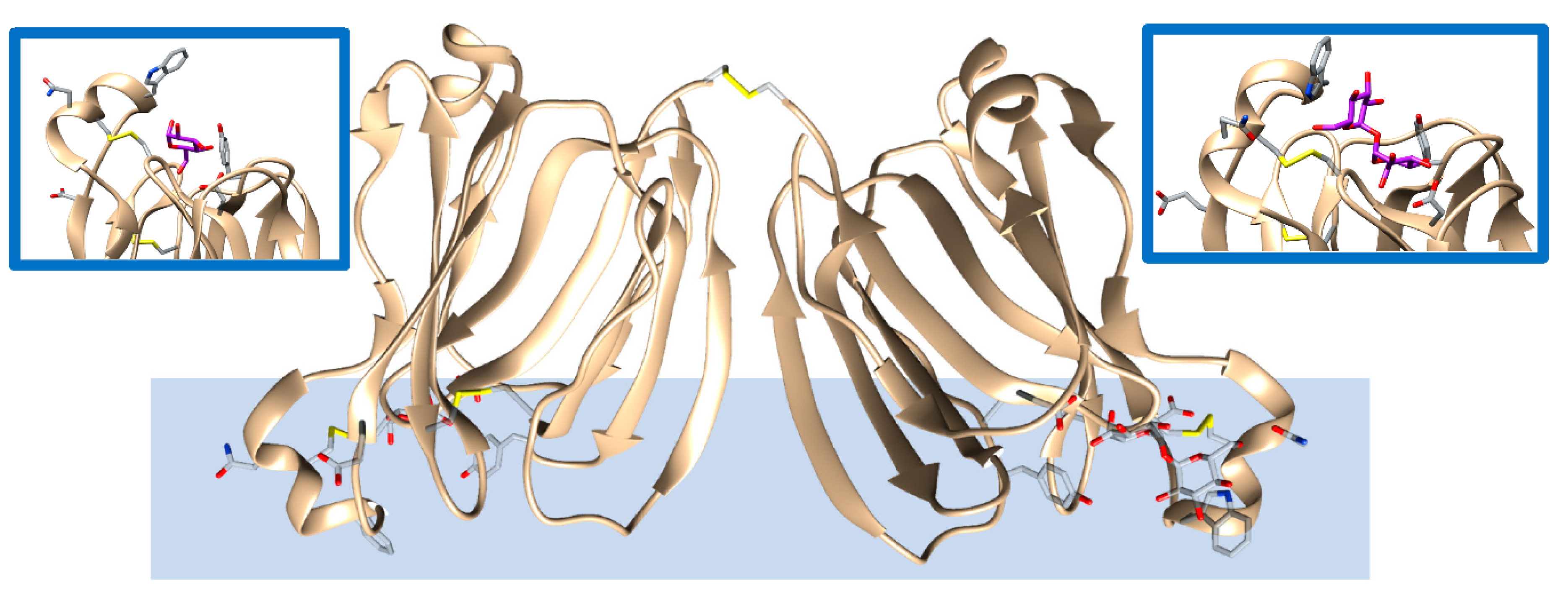
Figure 3. Lectin protein (PPL3A, brown, PDB entry 5yrf [50]) found in pearls and binding calcite biominerals. Amino acids and trehalose, in addition to the carbohydrate mediating the interactions with the biomineral, are highlithed as a stick representation. The biomineral is represented as a light blue color shape and is for rendition purposes only. Several intra- and one interchain -S-S bonds are present that could facilitate protein orientation for binding to the mineral surface. The insets provide zoom views of the bound carbohydrates (purple color) interacting with the represented side chains as found in the experimental structure.
2.2. Protein Templates and Biominerals in Plants
Biomineralization is also related to higher plants and biominerals play a variety of functions [80]. In fact, the precipitation of calcium oxalate within specialized plant cells has been associated with biochemically controlled processes and other cellular mechanisms [81]. L. Wang and co-workers have shown the presence of a nanofiber template protein that assists the formation of hundreds of aligned needle-shaped crystals called raphides (e.g., in banana) with a typical dimension of 150 μm in length and 4 μm in width [82]. This small protein (14 KDa) is conserved among a variety of plants and contains 16% calcium-binding amino acids of the total protein sequence, several of which include glutamate, aspartate, and cysteine. This protein has a C-terminal 11-mer proline rich peptide with a sequence (-RLPCPNCSKLP) that confers a helical shape similar to amelogenin, a binding mineral found in enamel [82,83][82][83]. In summary, the study of structural protein templates for biomineralization is not only limited to the formation of calcium carbonate or phosphate and the advancement of three-dimensional electron microscopy technique will allow imaging of new hybrid biomineral/protein superstructures at the atomic level [51].
References
- Veis, A. A Window on Biomineralization. Science 2005, 307, 1419–1420.
- Estroff, L.A. Introduction: Biomineralization. Chem. Rev. 2008, 108, 4329–4331.
- Sancetta, C. On biomineralization. Mar. Geol. 1993, 110, 184–185.
- Skinner, H.C.W.; Ehrlich, H. Biomineralization. In Treatise on Geochemistry, 2nd ed.; Elsevier: Oxford, UK, 2014; Volume 13, pp. 29–57. ISBN 978-0-08-098300-4.
- Ehrlich, H. Biocomposites and Mineralized Tissues. In Biological Materials of Marine Origin; Springer: Dordrecht, The Netherlands, 2014; pp. 91–210.
- Lowenstam, H.A. Minerals Formed by Organisms. Science 1981, 211, 1126–1131.
- Muller, W.E.G. Molecular Biomineralization: Aquatic Organisms forming Extraordinary Materials; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2011; ISBN 978-3-642-21229-1.
- Nakano, T.; Ikawa, N.; Ozimek, L. Chemical composition of chicken eggshell and shell membranes. Poult. Sci. 2003, 82, 510–514.
- Gautron, J.; Stapane, L.; Le Roy, N.; Nys, Y.; Rodriguez-Navarro, A.B.; Hincke, M.T. Avian eggshell biomineralization: An update on its structure, mineralogy and protein tool kit. BMC Mol. Cell Biol. 2021, 22, 11.
- Müller, F.D.; Schüler, D.; Pfeiffer, D. A Compass to Boost Navigation: Cell Biology of Bacterial Magnetotaxis. J. Bacteriol. 2020, 202, e00398-20.
- Rodríguez-Navarro, A.B.; Marie, P.; Nys, Y.; Hincke, M.T.; Gautron, J. Amorphous calcium carbonate controls avian eggshell mineralization: A new paradigm for understanding rapid eggshell calcification. J. Struct. Biol. 2015, 190, 291–303.
- Giesa, T.; Buehler, M.J. Nanoconfinement and the Strength of Biopolymers. Annu. Rev. Biophys. 2013, 42, 651–673.
- Kirschvink, J.L.; Walker, M.M.; Diebel, C.E. Magnetite-based magnetoreception. Curr. Opin. Neurobiol. 2001, 11, 462–467.
- Cadiou, H.; McNaughton, P. Avian magnetite-based magnetoreception: A physiologist’s perspective. J. R. Soc. Interface 2010, 7, S193–S205.
- Amor, M.; Ceballos, A.; Wan, J.; Simon, C.P.; Aron, A.T.; Chang, C.J.; Hellman, F.; Komeili, A. Magnetotactic Bacteria Accumulate a Large Pool of Iron Distinct from Their Magnetite Crystals. Appl. Environ. Microbiol. 2020, 86, e01278-20.
- Pósfai, M.; Kasama, T.; Dunin-Borkowski, R.E. Biominerals at the nanoscale: Biominerals at the nanoscale: Transmission electron microscopy methods for studying the special properties of biominerals. Eur. Mineral. Union Notes Mineral. 2013, 14, 377–435.
- McCausland, H.C.; Komeili, A. Magnetic genes: Studying the genetics of biomineralization in magnetotactic bacteria. PLoS Genet. 2020, 16, e1008499.
- Schüler, D. Formation of magnetosomes in magnetotactic bacteria. J. Mol. Microbiol. Biotechnol. 1999, 1, 79–86.
- Phoenix, V.R.; Konhauser, K.O. Benefits of bacterial biomineralization. Geobiology 2008, 6, 303–308.
- Dobro, M.J.; Oikonomou, C.M.; Piper, A.; Cohen, J.; Guo, K.; Jensen, T.; Tadayon, J.; Donermeyer, J.; Park, Y.; Solis, B.A.; et al. Uncharacterized Bacterial Structures Revealed by Electron Cryotomography. J. Bacteriol. 2017, 199, e00100-17.
- Monteil, C.L.; Vallenet, D.; Menguy, N.; Benzerara, K.; Barbe, V.; Fouteau, S.; Cruaud, C.; Floriani, M.; Viollier, E.; Adryanczyk, G.; et al. Ectosymbiotic bacteria at the origin of magnetoreception in a marine protist. Nat. Microbiol. 2019, 4, 1088–1095.
- Medakovi, D.; Popovi, S. Unusual Crystal Formation in Organisms-Exceptions that Confirm Biomineralization Rules. In Crystallization and Materials Science of Modern Artificial and Natural Crystals; BoD–Books on Demand: Nordstedt, Germany, 2012.
- Sugawara-Narutaki, A.; Nakamura, J.; Ohtsuki, C. Polymer-induced liquid precursors (PILPs) and bone regeneration. In Bioceramics; Elsevier: Amsterdam, The Netherlands, 2021; pp. 391–398.
- Knoll, A.H. Biomineralization and Evolutionary History. Rev. Miner. Geochem. 2003, 54, 329–356.
- Lefèvre, C.T.; Bazylinski, D.A. Ecology, Diversity, and Evolution of Magnetotactic Bacteria. Microbiol. Mol. Biol. Rev. 2013, 77, 497–526.
- Böhm, C.F.; Harris, J.; Schodder, P.I.; Wolf, S.E. Bioinspired Materials: From Living Systems to New Concepts in Materials Chemistry. Materials 2019, 12, 2117.
- Crichton, R. Biomineralization. In Biological Inorganic Chemistry; Elsevier: Amsterdam, The Netherlands, 2019.
- Cranford, S.W.; Buehler, M.J. The future of biomateriomics. In Biomateriomics; Springer: Dordrecht, The Netherlands, 2012; Volume 165.
- Harris, J.; Böhm, C.F.; Wolf, S.E. Universal structure motifs in biominerals: A lesson from nature for the efficient design of bioinspired functional materials. Interface Focus 2017, 7, 20160120.
- Vinn, O. The Role of Aragonite in Producing the Microstructural Diversity of Serpulid Skeletons. Minerals 2021, 11, 1435.
- Burley, S.K.; Bhikadiya, C.; Bi, C.; Bittrich, S.; Chen, L.; Crichlow, G.V.; Christie, C.H.; Dalenberg, K.; Di Costanzo, L.; Duarte, J.M.; et al. RCSB Protein Data Bank: Powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucleic Acids Res. 2020, 49, D437–D451.
- Boskey, A.L.; Villarreal-Ramirez, E. Intrinsically disordered proteins and biomineralization. Matrix Biol. 2016, 52, 43–59.
- Sodek, J.; Ganss, B.; McKee, M.D. Osteopontin. Crit. Rev. Oral Biol. Med. 2000, 11, 279–303.
- Davidov, G.; Abelya, G.; Zalk, R.; Izbicki, B.; Shaibi, S.; Spektor, L.; Shagidov, D.; Meyron-Holtz, E.G.; Zarivach, R.; Frank, G.A. Folding of an Intrinsically Disordered Iron-Binding Peptide in Response to Sedimentation Revealed by Cryo-EM. J. Am. Chem. Soc. 2020, 142, 19551–19557.
- Kim, I.W.; Collino, S.; Morse, A.D.E.; Evans, J.S. A Crystal Modulating Protein from Molluscan Nacre That Limits the Growth of Calcite In Vitro. Cryst. Growth Des. 2006, 6, 1078–1082.
- Collino, S.; Kim, I.W.; Evans, J.S. Identification and Structural Characterization of an Unusual RING-Like Sequence within an Extracellular Biomineralization Protein, AP7. Biochemistry 2008, 47, 3745–3755.
- Jackson, A.P.; Vincent, J.F.V.; Turner, R.M. The mechanical design of nacre. Proc. R. Soc. London B Biol. Sci. 1988, 234, 415–440.
- Chen, Y.; Feng, Y.; Deveaux, J.G.; Masoud, M.A.; Chandra, F.S.; Chen, H.; Zhang, D.; Feng, L. Biomineralization Forming Process and Bio-inspired Nanomaterials for Biomedical Application: A Review. Minerals 2019, 9, 68.
- Zhang, S.; Jiang, Z.; Shi, J.; Wang, X.; Han, P.; Qian, W. An Efficient, Recyclable, and Stable Immobilized Biocatalyst Based on Bioinspired Microcapsules-in-Hydrogel Scaffolds. ACS Appl. Mater. Interfaces 2016, 8, 25152–25161.
- Di Costanzo, L.; Ghosh, S.; Zardecki, C.; Burley, S.K. Using the Tools and Resources of the RCSB Protein Data Bank. Curr. Protoc. Bioinform. 2016, 55, 1.9.1–1.9.35.
- Jumper, J.; Evans, R.; Pritzel, A.; Green, T.; Figurnov, M.; Ronneberger, O.; Tunyasuvunakool, K.; Bates, R.; Žídek, A.; Potapenko, A.; et al. Highly accurate protein structure prediction with AlphaFold. Nature 2021, 596, 583–589.
- Hoang, Q.Q.; Sicheri, F.; Howard, A.J.; Yang, D.S.C. Bone recognition mechanism of porcine osteocalcin from crystal structure. Nature 2003, 425, 977–980.
- Dowd, T.L.; Rosen, J.F.; Li, A.L.; Gundberg, C.M. The Three-Dimensional Structure of Bovine Calcium Ion-Bound Osteocalcin Using 1H NMR Spectroscopy. Biochemistry 2003, 42, 7769–7779.
- Malashkevich, V.N.; Almo, S.C.; Dowd, T.L. X-ray Crystal Structure of Bovine 3 Glu-Osteocalcin. Biochemistry 2013, 52, 8387–8392.
- Frazão, C.; Simes, D.C.; Coelho, R.; Alves, D.; Williamson, M.K.; Price, P.A.; Cancela, M.L.; Carrondo, M.A. Structural Evidence of a Fourth Gla Residue in Fish Osteocalcin: Biological Implications. Biochemistry 2005, 44, 1234–1242.
- Ruiz-Arellano, R.R.; Medrano, F.J.; Moreno, A.; Romero, A. Structure of struthiocalcin-1, an intramineral protein from Struthio camelus eggshell, in two crystal forms. Acta Crystallogr. Sect. D Biol. Crystallogr. 2015, 71, 809–818.
- Reyes, J.P.; Moreno, A.; Romero, A. Crystal Structure of Ovocleidin-17, a Major Protein of the Calcified Gallus gallus Eggshell: Implications in the calcite mineral growth pattern. J. Biol. Chem. 2004, 279, 40876–40881.
- Sicheri, F.; Yang, D.S.C. Ice-binding structure and mechanism of an antifreeze protein from winter flounder. Nature 1995, 375, 427–431.
- Nishimiya, Y.; Kondo, H.; Takamichi, M.; Sugimoto, H.; Suzuki, M.; Miura, A.; Tsuda, S. Crystal Structure and Mutational Analysis of Ca2+-Independent Type II Antifreeze Protein from Longsnout Poacher, Brachyopsis rostratus. J. Mol. Biol. 2008, 382, 734–746.
- Nakae, S.; Shionyu, M.; Ogawa, T.; Shirai, T. Structures of jacalin-related lectin PPL3 regulating pearl shell biomineralization. Proteins Struct. Funct. Bioinform. 2018, 86, 644–653.
- Görlich, S.; Samuel, A.J.; Best, R.J.; Seidel, R.; Vacelet, J.; Leonarski, F.K.; Tomizaki, T.; Rellinghaus, B.; Pohl, D.; Zlotnikov, I. Natural hybrid silica/protein superstructure at atomic resolution. Proc. Natl. Acad. Sci. USA 2020, 117, 31088–31093.
- Fairhead, M.; Johnson, K.A.; Kowatz, T.; McMahon, S.A.; Carter, L.G.; Oke, M.; Liu, H.; Naismith, J.H.; van der Walle, C.F. Crystal structure and silica condensing activities of silicatein α–cathepsin L chimeras. Chem. Commun. 2008, 15, 1765–1767.
- Zeytuni, N.; Ozyamak, E.; Ben-Harush, K.; Davidov, G.; Levin, M.; Gat, Y.; Moyal, T.; Brik, A.; Komeili, A.; Zarivach, R. Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly. Proc. Natl. Acad. Sci. USA 2011, 108, E480–E487.
- Uebe, R.; Keren-Khadmy, N.; Zeytuni, N.; Katzmann, E.; Navon, Y.; Davidov, G.; Bitton, R.; Plitzko, J.M.; Schüler, D.; Zarivach, R. The dual role of MamB in magnetosome membrane assembly and magnetite biomineralization. Mol. Microbiol. 2018, 107, 542–557.
- Quinlan, A.; Murat, D.; Vali, H.; Komeili, A. The HtrA/DegP family protease MamE is a bifunctional protein with roles in magnetosome protein localization and magnetite biomineralization. Mol. Microbiol. 2011, 80, 1075–1087.
- Zeytuni, N.; Uebe, R.; Maes, M.; Davidov, G.; Baram, M.; Raschdorf, O.; Nadav-Tsubery, M.; Kolusheva, S.; Bitton, R.; Goobes, G.; et al. Cation Diffusion Facilitators Transport Initiation and Regulation Is Mediated by Cation Induced Conformational Changes of the Cytoplasmic Domain. PLoS ONE 2014, 9, e92141.
- Bergeron, J.R.; Hutto, R.; Ozyamak, E.; Hom, N.; Hansen, J.; Draper, O.; Byrne, M.E.; Keyhani, S.; Komeili, A.; Kollman, J.M. Structure of the magnetosome-associated actin-like MamK filament at subnanometer resolution. Protein Sci. 2016, 26, 93–102.
- Hershey, D.M.; Ren, X.; Melnyk, R.A.; Browne, P.J.; Ozyamak, E.; Jones, S.R.; Chang, M.C.Y.; Hurley, J.H.; Komeili, A. MamO Is a Repurposed Serine Protease that Promotes Magnetite Biomineralization through Direct Transition Metal Binding in Magnetotactic Bacteria. PLOS Biol. 2016, 14, e1002402.
- Siponen, M.I.; Legrand, P.; Widdrat, M.; Jones, S.R.; Zhang, W.-J.; Chang, M.C.Y.; Faivre, D.; Arnoux, P.; Pignol, D. Structural insight into magnetochrome-mediated magnetite biomineralization. Nature 2013, 502, 681–684.
- Voet, A.R.D.; Noguchi, H.; Addy, C.; Zhang, K.Y.J.; Tame, J.R.H. Biomineralization of a Cadmium Chloride Nanocrystal by a Designed Symmetrical Protein. Angew. Chem. Int. Ed. 2015, 54, 9857–9860.
- Wang, Y.; Chen, H.; Huang, Z.; Yang, M.; Yu, H.; Peng, M.; Yang, Z.; Chen, S. Structural characterization of cystathionine γ-lyase smCSE enables aqueous metal quantum dot biosynthesis. Int. J. Biol. Macromol. 2021, 174, 42–51.
- Thaker, A.; Sirajudeen, L.; Simmons, C.R.; Nannenga, B.L. Structure-guided identification of a peptide for bio-enabled gold nanoparticle synthesis. Biotechnol. Bioeng. 2021, 118, 4867–4873.
- Wei, H.; Wang, Z.; Zhang, J.; House, S.; Gao, Y.-G.; Yang, L.; Robinson, H.; Tan, L.H.; Xing, H.; Hou, C.; et al. Time-dependent, protein-directed growth of gold nanoparticles within a single crystal of lysozyme. Nat. Nanotechnol. 2011, 6, 93–97.
- Butts, C.A.; Swift, J.; Kang, S.-G.; Di Costanzo, L.; Christianson, D.W.; Saven, J.G.; Dmochowski, I.J. Directing Noble Metal Ion Chemistry within a Designed Ferritin Protein. Biochemistry 2008, 47, 12729–12739.
- Kasyutich, O.; Ilari, A.; Fiorillo, A.; Tatchev, D.; Hoell, A.; Ceci, P. Silver Ion Incorporation and Nanoparticle Formation inside the Cavity of Pyrococcus furiosus Ferritin: Structural and Size-Distribution Analyses. J. Am. Chem. Soc. 2010, 132, 3621–3627.
- Tosha, T.; Ng, H.-L.; Bhattasali, O.; Alber, T.; Theil, E.C. Moving Metal Ions through Ferritin-Protein Nanocages from Three-Fold Pores to Catalytic Sites. J. Am. Chem. Soc. 2010, 132, 14562–14569.
- Zoch, M.L.; Clemens, T.L.; Riddle, R.C. New insights into the biology of osteocalcin. Bone 2015, 82, 42–49.
- Rehder, D.S.; Gundberg, C.M.; Booth, S.L.; Borges, C.R. Gamma-Carboxylation and Fragmentation of Osteocalcin in Human Serum Defined by Mass Spectrometry. Mol. Cell. Proteom. 2015, 14, 1546–1555.
- Reyes-Grajeda, J.; Jauregui-Zuniga, D.; Rodriguez-Romero, A.; Hernandez-Santoyo, A.; Bolanos-Garcia, V.; Moreno, A. Crystallization and Preliminary X-ray Analysis of Ovocleidin-17 A Major Protein of the Gallus Gallus Eggshell Calcified Layer. Protein Pept. Lett. 2002, 9, 253–257.
- Zardecki, C.; Dutta, S.; Goodsell, D.S.; Lowe, R.; Voigt, M.; Burley, S.K. PDB-101: Educational resources supporting molecular explorations through biology and medicine. Protein Sci. 2021, 31, 129–140.
- Lakshminarayanan, R.; Valiyaveettil, S.; Rao, V.S.; Kini, M. Purification, Characterization, and in VitroMineralization Studies of a Novel Goose Eggshell Matrix Protein, Ansocalcin. J. Biol. Chem. 2003, 278, 2928–2936.
- Lakshminarayanan, R.; Kini, R.M.; Valiyaveettil, S. Investigation of the role of ansocalcin in the biomineralization in goose eggshell matrix. Proc. Natl. Acad. Sci. USA 2002, 99, 5155–5159.
- Demarchi, B.; Hall, S.; Roncal-Herrero, T.; Freeman, C.L.; Woolley, J.; Crisp, M.K.; Wilson, J.; Fotakis, A.; Fischer, R.; Kessler, B.M.; et al. Protein sequences bound to mineral surfaces persist into deep time. eLife 2016, 5, e17092.
- Saitta, E.T.; Vinther, J.; Crisp, M.K.; Abbott, G.D.; Kaye, T.G.; Pittman, M.; Bull, I.; Fletcher, I.; Chen, X.; Collins, M.J.; et al. Non-avian dinosaur eggshell calcite contains ancient, endogenous amino acids. bioRxiv 2020.
- Gharib, G.; Saeidiharzand, S.; Sadaghiani, A.K.; Koşar, A. Antifreeze Proteins: A Tale of Evolution from Origin to Energy Applications. Front. Bioeng. Biotechnol. 2022, 9, 770588.
- Zhang, G.; Fang, X.; Guo, X.; Li, L.; Luo, R.; Xu, F.; Yang, P.; Zhang, L.; Wang, X.; Qi, H.; et al. The oyster genome reveals stress adaptation and complexity of shell formation. Nature 2012, 490, 49–54.
- Ginebra, M.-P.; Espanol, M.; Maazouz, Y.; Bergez, V.; Pastorino, D. Bioceramics and bone healing. EFORT Open Rev. 2018, 3, 173–183.
- Dhami, N.K.; Reddy, M.S.; Mukherjee, M.S. Biomineralization of calcium carbonates and their engineered applications: A review. Front. Microbiol. 2013, 4, 314.
- Pei, J.; Wang, Y.; Zou, X.; Ruan, H.; Tang, C.; Liao, J.; Si, G.; Sun, P. Extraction, Purification, Bioactivities and Application of Matrix Proteins From Pearl Powder and Nacre Powder: A Review. Front. Bioeng. Biotechnol. 2021, 9, 649665.
- He, H.; Veneklaas, E.J.; Kuo, J.; Lambers, H. Physiological and ecological significance of biomineralization in plants. Trends Plant Sci. 2014, 19, 166–174.
- Weiner, S.; Addadi, L. Crystallization Pathways in Biomineralization. Annu. Rev. Mater. Sci. 2011, 41, 21–40.
- Li, X.; Zhang, W.; Lu, J.; Huang, L.; Nan, D.; Webb, M.A.; Hillion, F.; Wang, L. Templated Biomineralization on Self-Assembled Protein Nanofibers Buried in Calcium Oxalate Raphides of Musa spp. Chem. Mater. 2014, 26, 3862–3869.
- Friddle, R.W.; Battle, K.; Trubetskoy, V.; Tao, J.; Salter, E.A.; Moradian-Oldak, J.; De Yoreo, J.J.; Wierzbicki, A. Single-molecule determination of the face-specific adsorption of Amelogenin’s C-terminus on hydroxyapatite. Angew. Chem. Int. Ed. 2011, 50, 7541–7545.
More
