Embryonic development is a highly intricate and complex process. Different regulatory mechanisms cooperatively dictate the fate of cells as they progress from pluripotent stem cells to terminally differentiated cell types in tissues. A crucial regulator of these processes is the Polycomb Repressive Complex 2 (PRC2). By catalyzing the mono-, di-, and tri-methylation of lysine residues on histone H3 tails (H3K27me3), PRC2 compacts chromatin by cooperating with Polycomb Repressive Complex 1 (PRC1) and represses transcription of target genes. Proteomic and biochemical studies have revealed two variant complexes of PRC2, namely PRC2.1 which consists of the core proteins (EZH2, SUZ12, EED, and RBBP4/7) interacting with one of the Polycomb-like proteins (MTF2, PHF1, PHF19), and EPOP or PALI1/2, and PRC2.2 which contains JARID2 and AEBP2 proteins. MTF2 and JARID2 have been discovered to have crucial roles in directing and recruiting PRC2 to target genes for repression in embryonic stem cells (ESCs).
- Polycomb
- H3K27me3
- H2AK119ub
- pluripotency
- embryogenesis
- lineage commitment
1. Introduction
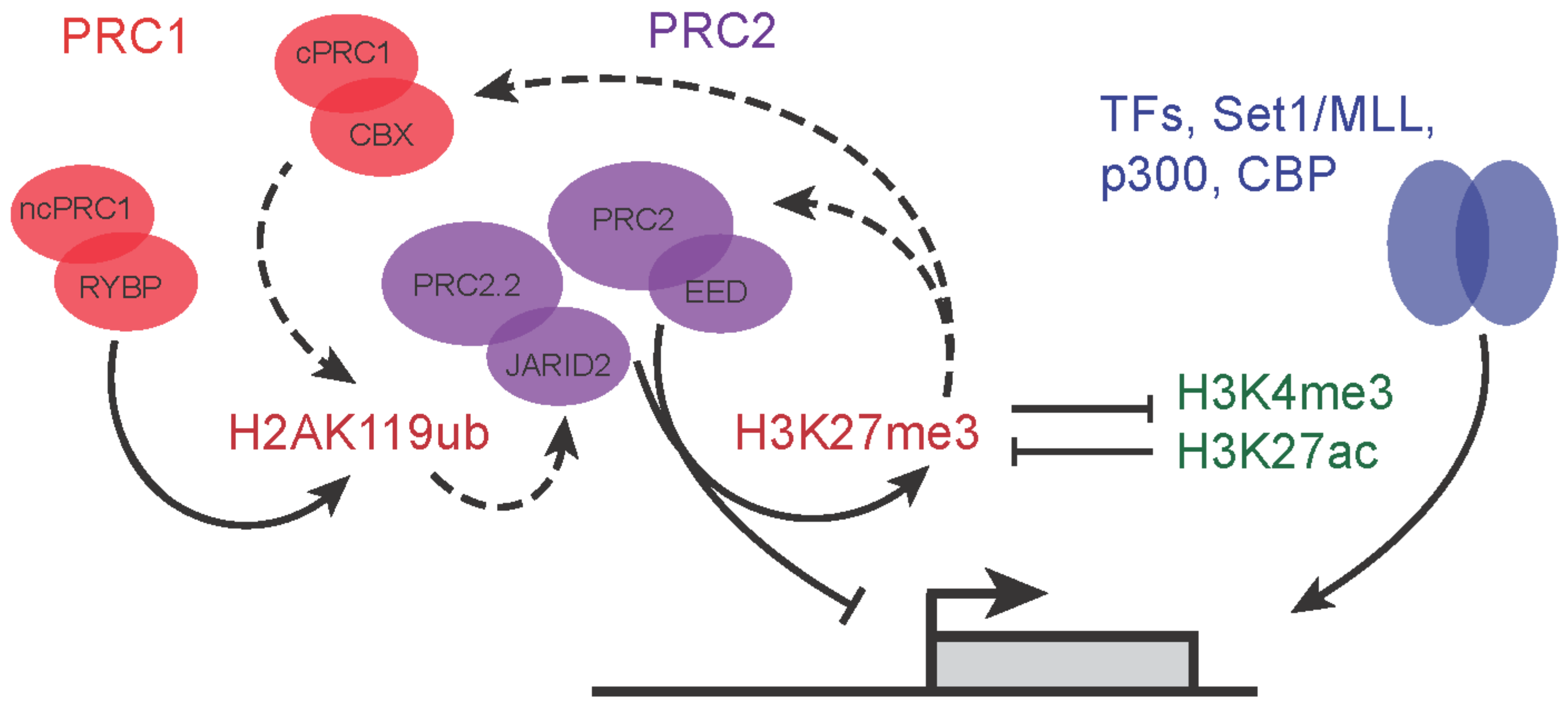
2. More Than the Sum of Its Parts—Polycomb Proteins and Their Molecular Functions
The biochemical properties of Polycomb complexes have important implications for their roles in development. Biochemically distinct variant complexes have been described for both PRC1 and PRC2 (Figure 1). PRC1 is made up of at least eight different variant forms and they are classified based on different interactors with the E3 Ligase RING1A/B (RING1, RNF2) [1]. Canonical PRC1 (cPRC1) consists of RING1A/B with SCMH and PHC proteins which are associated with chromatin compaction, PCGF2/4 proteins that ensure the stabilization and integrity of the complex, and CBX proteins which both compact chromatin and aid in targeting of PRC1 to genomic loci [24,25,26,27,28][24][25][26][27][28]. Non-canonical PRC1, on the other hand, contains either the RING1 and YY1 binding protein (RYBP) or YY1-associated factor 2 (YAF2) protein associating with RING1A/B and different variants of the PCGF proteins [29,30,31][29][30][31]. For example, the PRC1.1 variant complex comprises the ncPRC1 core consisting of RING1A/B and RYBP/YAF2, together with PCGF1 and KDM2B. KDMB2B is crucial in targeting RING1B to unmethylated CpG islands. Functionally, key differences between cPRC1 and ncPRC1 include their ability to interact with H3K27me3 via the chromodomains found in CBX proteins of cPRC1 [12], and the much lower ubiquitin ligase activity of cPRC1 compared to ncPRC1 complexes. The interaction of CBX proteins of cPRC1 with H3K27me3 underscores the potential for functional interactions with PRC2 activity [32]. ncPRC1 complexes have other mechanisms of recruitment that are independent of PRC2 and H3K27me3 [31,33][31][33].
3. Roles of Polycomb Group Proteins in Pluripotency and Early Development
Based on the roles of Polycomb in homeobox gene regulation in D. melanogaster development [50[47][48][49][50],51,52,53], it is no surprise that the Polycomb proteins serve important roles in mammalian development. Early loss-of-function studies in mice have outlined the critical importance of core Polycomb proteins, particularly in early developmental processes such as gastrulation and body-axis formation [17,54,55,56][17][51][52][53]. Although the studies were conducted using various loss-of-function methods, the picture emerged that embryos could not survive past gastrulation, without the PRC2 core proteins [17,55,56][17][52][53]. Recent studies identified an important role for PRC2 in restricting trophoblast induction in human naïve pluripotent stem cells in vitro [57[54][55],58], suggesting a role in the earliest mammalian lineage decision after fertilization. In PRC1, the loss of RING1B protein results in developmental arrest at E6.5, during gastrulation, but RING1A mutants were able to survive and reach birth, although displaying defects in transformation of the axial skeleton and altered levels of Hox gene expression [59,60][56][57]. On the other hand, the loss of some other non-core subunits, such as the PCLs and JARID2/AEBP2 of PRC2 in mice, revealed a later embryonic lethality [23,61][23][58]. Though these subunits are essential for normal development, this could suggest that their role in overall PRC2 activity is more subtle, is buffered by other activities at earlier stages, or is more selectively required for specific cell lineages or developmental processes. Their roles may also be contextualized by the expression levels of both PRC2 and the activators they antagonize in specific cell lineages and stages of differentiation. Table 1 provides an overview of the Polycomb loss-of-function mutations during development, outlining the different cell-types or lineages that the Polycomb proteins were studied in.| Complex | Variant Complex | Non Core Subunit | In Vivo Developmental Phenotype | In Vitro Differentiation Phenotype | Cell Type or Tissue Specificity | References |
|---|---|---|---|---|---|---|
| PRC2 | PRC2.1 | MTF2 | Embryonic lethal due to severe anemia around embryonic day 15.5 | Mtf2 null cells result in faster differentiation towards all cell types in EBs | ESC/Gastrula | Faust et al., 1995: Hojfeldt et al., 2018; Perino et al.,2018; Loh et al., 2021; Rothberg et al., 2018 |
| PHF1 | ND | Increased stoichometry relative to EED during NPC differentiation | Neuronal Precursors | Kloet et al., 2016 | ||
| PHF19 | ND | Enhanced erythrocyte differentiation | Mesodermal | García-Montolio et al., 2021 | ||
| EPOP | ND | ND | ND | |||
| PALI1/2 | Lethal between E11.5 and perinatal lethality | Deregulation and reduction of H3K27me3 on PRC2 target genes | ND | Conway, E. et al., 2018 | ||
| PRC2.2 | JARID2 | Lethal between E10.5 - E18.5 | Stagnated differentiation towards all germ layers | ND | Loh et al., 2021 | |
| AEBP2 | Late-embryonic and perinatal lethality; skeletal transformations | Deregulation and increase of H3K27me3 on PRC2 target genes | ND | Grijzenhout, A. et al., 2016; Kim, H. et al., 2011 |
||
| cPRC1 | CBX2 | Male to female sex reversal | Activator of the testis - determining gene Sry |
Testes, Sex Organ | Katoh-Fukui, Y. et al., 1998 | |
| CBX4 | Pre-weaning lethality | ND | ND | |||
| CBX6 | Several transformational defects in late embryonic development | ND | Enriched in NPC differentiation | Dickinson, M. E. et al., 2016 | ||
| CBX7 | ND | Enriched in early ESC stages | ||||
| CBX8 | ND | ND | Enriched in NPC differentiation | Morey, L. et al. 2012 | ||
| PHC1 | Perinatal lethality | Destabilized pluripotency by disruption of Nanog long-range interactions |
ND | Isono K. et al., 2005 | ||
| PHC2 | Posterior transformation; Hox gene deregulation; Viable birth | ND | ND | Isono K. et al., 2005 | ||
| PHC3 | Phc3 null animals are born but exhibit cardiac abnormalities | ND | ND | Dickinson, M. E. et al., 2016 |
||
| PCGF2 | Posterior transformation; Hox gene deregulation; Viable birth | ND | ND | Akasaka, T. et al., 1996 | ||
| PCGF4 | Perinatal lethality, ataxia, severe haematopoietic defects | ND | ND | van der Lugt, N. M. et al., 1994 |
||
| ncPRC1 | CBX3 | Defects in germ cell development | Inhibition results in neural differentiation impairment and diverting towards mesendodermal differentiion |
Neural Precursors | Abe, K. et al., 2011 | |
| PCGF1 | E12.5 lethality | Promotes ectoderm and mesoderm differentiation | Ectoderm and Mesoderm lineages | Yan Y et al., 2017 | ||
| PCGF3 | Double Pcgf3;Pcgf5 deletion results in female-specific embryonic lethality | ND | ND | Almeida, M. et al., 2017 | ||
| PCGF5 | ||||||
| PCGF6 | defects in pre-implantation and periimplantation and in placenta development | Ablation of Pcgf6 in ESCs leads to robust de-repression of such germ cell-related genes, in turn affecting cell growth and viability. | pre- and peri-implantation mouse embryo | Dickinson, M. E. et al., 2016 |
||
| KDM2B | Mid-gestation lethality; posterior transformation of the axial skeleton | Maintain pluripotency by recruitment of PRC1 to CpGs | ND | Boulard, M. et al., 2015 | ||
| RYBP | E6.5 Lethality; impaired cell proliferation | Depletion of Rybp inhibits proliferation and promotes Neuronal differentiation of embryonic neural progenitor cells (eNPCs) |
Neural Precursors | Pirity, M. K. et al., 2015 | ||
| AUTS2 | Die before weaning with defects in the nervous, cardiovascular and biliary systems |
ND | Neural Ectoderm | Gao, Z. et al., 2014 | ||
| L3MBTL2 | E6.5 lethality; gastrulation defects | Compromised proliferation and abnormal differentiation of L3mbtl2(-/) embryonic stem (ES) cells | ESC | Qin, J. et al., 2012 |
3.1. PRC2 Variant Complexes and Their Roles in Embryonic Stem Cells
In terms of PRC2, several major landmark papers uncovered the mechanism by which MTF2 (PRC2.1) can directly and selectively bind to a subset of unmethylated CpG-dense DNA regions, with preferential binding to sites with specific DNA shape properties such as an underwound helix as compared to the canonical B-DNA structure [36,37][36][37]. This finding was supported by crystal structures of the PCL proteins MTF2 and PHF1 bound to DNA, demonstrating a binding of the winged-helix domain to DNA with unmethylated CpGs and reduced helical twist [37]. The loss of MTF2 in mESCs resulted in globally reduced H3K27me3 levels, and recruitment of EZH2 was also severely reduced at target gene locations [36,45,46,62][36][45][46][59]. Unlike MTF2, the loss of other PCL proteins, PHF1 and PHF19 did not elicit a similar level of drastic reduction of PRC2 recruitment in mESCs [45,62][45][59]. This might be because these proteins are expressed at much lower levels compared to MTF2 in mESCs [63][60]. The other PRC2.1 protein that plays an important role in mESCs is EPOP. Deletion of EPOP in mESCs prevents PRC2.1 from interacting with Elongin B/C proteins, which in turn reduces RNA polymerase II elongation on target genes [64][61]. Therefore, while the loss of EPOP does not directly translate into loss of H3K27me3 levels, it would set up for a low level of permissive transcription at target genes and affect how PRC2 regulates transcription. The interaction of EPOP with PRC2 is found to be mutually exclusive with PALI1/2 [38,40][38][40]. This is corroborated by studies demonstrating a dynamic shift of the PRC2.1 interactome between EPOP and PALI1/2 during mESC differentiation [63][60]. Unlike EPOP, the ablation of PALI1/2 proteins resulted in some degree of H3K27me3 reduction at target genes [39]. This indicates the importance of these proteins in maintaining the catalytic function of PRC2, but just like PHF1 and PHF19, their stoichiometry relative to other PRC2.1 proteins is found to be minimal.4. Roles of Polycomb Complexes in Developmental Cell Lineages
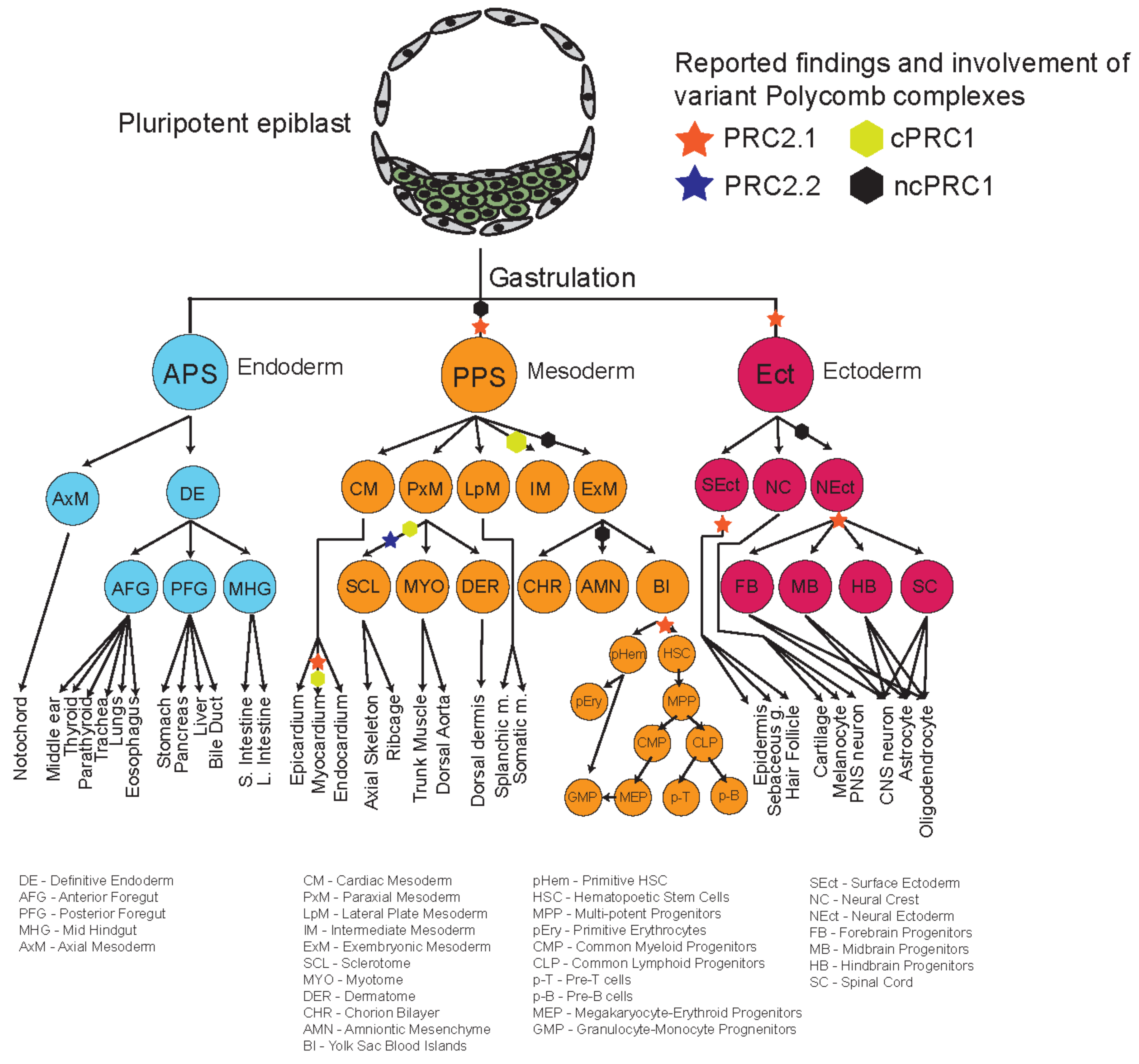
The repertoire of different phenotypes observed for many mutants of different Polycomb proteins, and their relatively delayed phenotypes suggest that some of these variant complex subunits might play more important roles during organogenesis and later developmental stages (Figure 3). Recent studies looking into the roles of different Polycomb proteins during in vitro differentiation shed some light on the potential specific mechanisms and roles of these different proteins during development. Figure 4 provides an overview and developmental roadmap of the known contributions of Polycomb complexes during development. It is apparent that there are still many developmental cell types, particularly from endoderm lineages, that remain to be investigated for contributions of Polycomb complexes.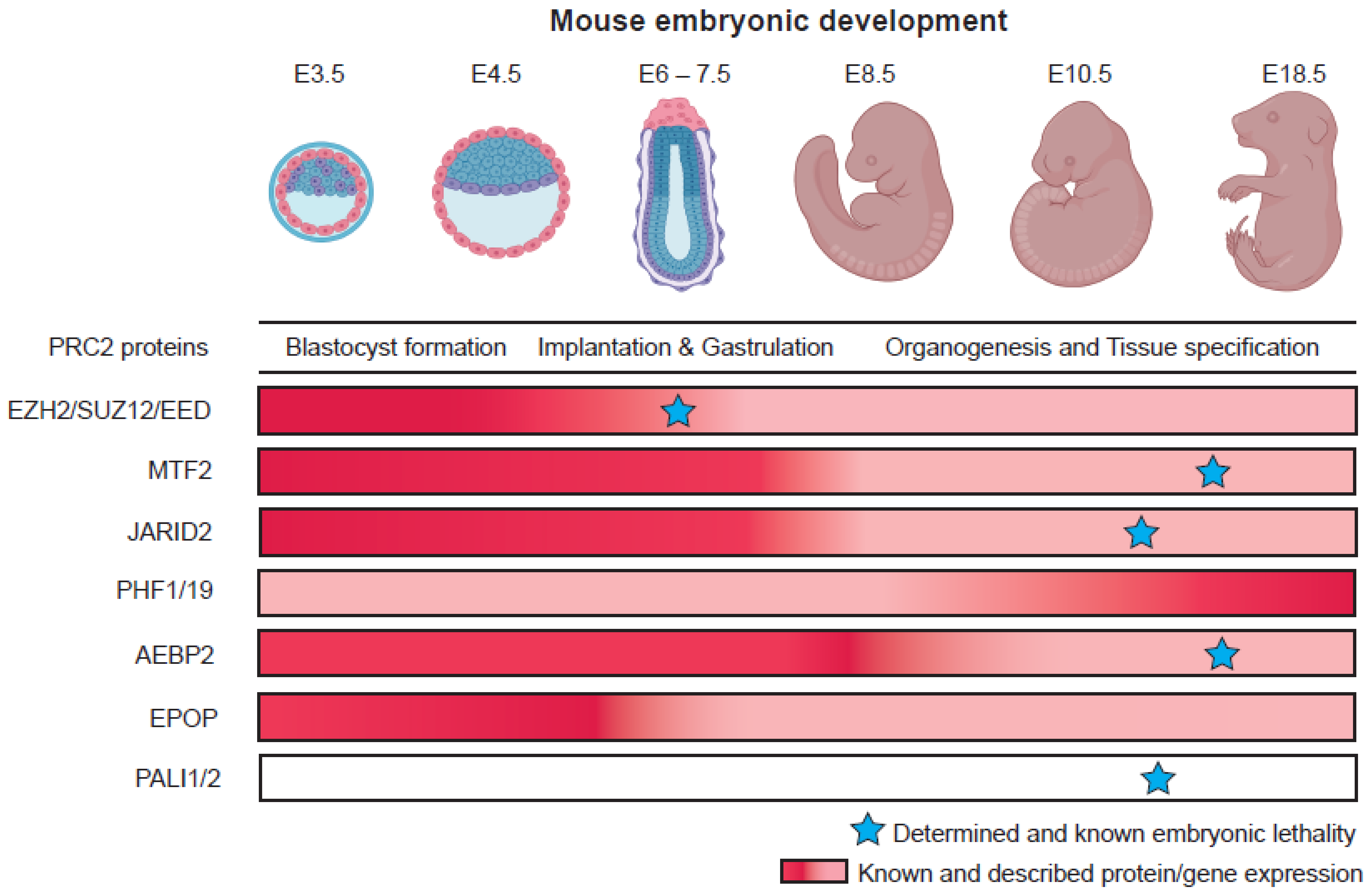
References
- Wang, H.; Wang, L.; Erdjument-Bromage, H.; Vidal, M.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H2A ubiquitination in Polycomb silencing. Nature 2004, 431, 873–878.
- Schuettengruber, B.; Cavalli, G. Recruitment of Polycomb group complexes and their role in the dynamic regulation of cell fate choice. Development 2009, 136, 3531–3542.
- Simon, J.A.; Kingston, R.E. Mechanisms of Polycomb gene silencing: Knowns and unknowns. Nat. Rev. Mol. Cell Biol. 2009, 10, 697–708.
- Van Mierlo, G.; Veenstra, G.J.C.; Vermeulen, M.; Marks, H. The Complexity of PRC2 Subcomplexes. Trends Cell Biol. 2019, 29, 660–671.
- Cao, R.; Wang, L.; Wang, H.; Xia, L.; Erdjument-Bromage, H.; Tempst, P.; Jones, R.S.; Zhang, Y. Role of histone H3 lysine 27 methylation in polycomb-group silencing. Science 2002, 298, 1039–1043.
- Müller, J.; Hart, C.M.; Francis, N.J.; Vargas, M.L.; Sengupta, A.; Wild, B.; Miller, E.L.; O’Connor, M.B.; Kingston, R.E.; Simon, J.A. Histone Methyltransferase Activity of a Drosophila Polycomb Group Repressor Complex. Cell 2002, 111, 197–208.
- Blackledge, N.P.; Fursova, N.A.; Kelley, J.R.; Huseyin, M.K.; Feldmann, A.; Klose, R.J. PRC1 Catalytic Activity Is Central to Polycomb System Function. Mol. Cell 2020, 77, 857–874.
- Loh, C.H.; van Genesen, S.; Perino, M.; Bark, M.R.; Veenstra, G.J.C. Loss of PRC2 subunits primes lineage choice during exit of pluripotency. Nat. Commun. 2021, 12, 1–14.
- Cooper, S.; Grijzenhout, A.; Underwood, E.; Ancelin, K.; Zhang, T.; Nesterova, T.B.; Anil-Kirmizitas, B.; Bassett, A.; Kooistra, S.M.; Agger, K.; et al. Jarid2 binds mono-ubiquitylated H2A lysine 119 to mediate crosstalk between Polycomb complexes PRC1 and PRC2. Nat. Commun. 2016, 7, 13661.
- Kalb, R.; Latwiel, S.; Baymaz, H.I.; Jansen, P.W.; Müller, C.W.; Vermeulen, M.; Müller, J. Histone H2A monoubiquitination promotes histone H3 methylation in Polycomb repression. Nat. Struct. Mol. Biol. 2014, 21, 569–571.
- Tamburri, S.; Lavarone, E.; Fernández-Pérez, D.; Conway, E.; Zanotti, M.; Manganaro, D.; Pasini, D. Histone H2AK119 Mono-Ubiquitination Is Essential for Polycomb-Mediated Transcriptional Repression. Mol. Cell 2020, 77, 840–856.
- Tavares, L.; Dimitrova, E.; Oxley, D.; Webster, J.; Poot, R.; Demmers, J.; Bezstarosti, K.; Taylor, S.; Ura, H.; Koide, H.; et al. RYBP-PRC1 complexes mediate H2A ubiquitylation at polycomb target sites independently of PRC2 and H3K27me3. Cell 2012, 148, 664–678.
- Morey, L.; Helin, K. Polycomb group protein-mediated repression of transcription. Trends Biochem. Sci. 2010, 35, 323–332.
- Piunti, A.; Shilatifard, A. Epigenetic balance of gene expression by polycomb and compass families. Science 2016, 352, aad9780.
- Boulet, A.M.; Capecchi, M.R. Signaling by FGF4 and FGF8 is required for axial elongation of the mouse embryo. Dev. Biol. 2012, 371, 235–245.
- Pasini, D.; Bracken, A.P.; Jensen, M.R.; Denchi, E.L.; Helin, K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004, 23, 4061–4071.
- O’Carroll, D.; Erhardt, S.; Pagani, M.; Barton, S.C.; Surani, M.A.; Jenuwein, T. The Polycomb-Group Gene Ezh2 Is Required for Early Mouse Development. Mol. Cell. Biol. 2001, 21, 4330–4336.
- Rothberg, J.L.M.; Maganti, H.B.; Jrade, H.; Porter, C.J.; Palidwor, G.A.; Cafariello, C.; Battaion, H.L.; Khan, S.T.; Perkins, T.J.; Paulson, R.F.; et al. Mtf2-PRC2 control of canonical Wnt signaling is required for definitive erythropoiesis. Cell Discov. 2018, 4, 21.
- Conway, E.; Healy, E.; Bracken, A.P. PRC2 mediated H3K27 methylations in cellular identity and cancer. Curr. Opin. Cell Biol. 2015, 37, 42–48.
- Deevy, O.; Bracken, A.P. PRC2 functions in development and congenital disorders. Dev. Camb 2019, 146, dev181354.
- Li, X.; Isono, K.; Yamada, D.; Endo, T.A.; Endoh, M.; Shinga, J.; Mizutani-Koseki, Y.; Otte, A.P.; Casanova, M.; Kitamura, H.; et al. Mammalian Polycomb-Like Pcl2/Mtf2 Is a Novel Regulatory Component of PRC2 That Can Differentially Modulate Polycomb Activity both at the Hox Gene Cluster and at Cdkn2a Genes. Mol. Cell. Biol. 2011, 31, 351–364.
- Grijzenhout, A.; Godwin, J.; Koseki, H.; Gdula, M.R.; Szumska, D.; McGouran, J.F.; Bhattacharya, S.; Kessler, B.M.; Brockdorff, N.; Cooper, S. Functional analysis of AEBP2, a PRC2 polycomb protein, reveals a trithorax phenotype in embryonic development and in ESCs. Dev. Camb. 2016, 143, 2716–2723.
- Kim, H.; Kang, K.; Ekram, M.B.; Roh, T.Y.; Kim, J. Aebp2 as an epigenetic regulator for neural crest cells. PLoS ONE 2011, 6, e25174.
- Wang, R.; Taylor, A.B.; Leal, B.Z.; Chadwell, L.V.; Ilangovan, U.; Robinson, A.K.; Schirf, V.; Hart, P.J.; Lafer, E.M.; Demeler, B.; et al. Polycomb group targeting through different binding partners of RING1B C-terminal domain. Structure 2010, 18, 966–975.
- Messmer, S.; Franke, A.; Paro, R. Analysis of the functional role of the Polycomb chromo domain in Drosophila melanogaster. Genes Dev. 1992, 6, 1241–1254.
- Levine, S.S.; Weiss, A.; Erdjument-Bromage, H.; Shao, Z.; Tempst, P.; Kingston, R.E. The Core of the Polycomb Repressive Complex Is Compositionally and Functionally Conserved in Flies and Humans. Mol. Cell. Biol. 2002, 22, 6070–6078.
- Scelfo, A.; Fernández-Pérez, D.; Tamburri, S.; Zanotti, M.; Lavarone, E.; Soldi, M.; Bonaldi, T.; Ferrari, K.J.; Pasini, D. Functional Landscape of PCGF Proteins Reveals Both RING1A/B-Dependent-and RING1A/B-Independent-Specific Activities. Mol. Cell 2019, 74, 1037–1052.
- Fursova, N.A.; Blackledge, N.P.; Nakayama, M.; Ito, S.; Koseki, Y.; Farcas, A.M.; King, H.W.; Koseki, H.; Klose, R.J. Synergy between Variant PRC1 Complexes Defines Polycomb-Mediated Gene Repression. Mol. Cell 2019, 74, 1020–1036.
- Garcia, E.; Marcos-Gutiérrez, C.; Del Mar Lorente, M.; Moreno, J.C.; Vidal, M. RYBP, a new repressor protein that interacts with components of the mammalian Polycomb complex, and with the transcription factor YY1. EMBO J. 1999, 18, 3404–3418.
- Arrigoni, R.; Alam, S.L.; Wamstad, J.A.; Bardwell, V.J.; Sundquist, W.I.; Schreiber-Agus, N. The Polycomb-associated protein Rybp is a ubiquitin binding protein. FEBS Lett. 2006, 580, 6233–6241.
- Morey, L.; Aloia, L.; Cozzuto, L.; Benitah, S.A.; Di Croce, L. RYBP and Cbx7 Define Specific Biological Functions of Polycomb Complexes in Mouse Embryonic Stem Cells. Cell Rep. 2013, 3, 60–69.
- Laugesen, A.; Højfeldt, J.W.; Helin, K. Molecular Mechanisms Directing PRC2 Recruitment and H3K27 Methylation. Mol. Cell 2019, 74, 8–18.
- Rose, N.R.; King, H.W.; Blackledge, N.P.; Fursova, N.A.; Ember, K.J.; Fischer, R.; Kessler, B.M.; Klose, R.J. RYBP stimulates PRC1 to shape chromatin-based communication between polycomb repressive complexes. eLife 2016, 5, e18591.
- Czermin, B.; Melfi, R.; McCabe, D.; Seitz, V.; Imhof, A.; Pirrotta, V. Drosophila enhancer of Zeste/ESC complexes have a histone H3 methyltransferase activity that marks chromosomal Polycomb sites. Cell 2002, 111, 185–196.
- Kuzmichev, A.; Nishioka, K.; Erdjument-Bromage, H.; Tempst, P.; Reinberg, D. Histone methyltransferase activity associated with a human multiprotein complex containing the enhancer of zeste protein. Genes Dev. 2002, 16, 2893–2905.
- Perino, M.; van Mierlo, G.; Karemaker, I.D.; van Genesen, S.; Vermeulen, M.; Marks, H.; van Heeringen, S.J.; Veenstra, G.J.C. MTF2 recruits Polycomb Repressive Complex 2 by helical-shape-selective DNA binding. Nat. Genet. 2018, 50, 1002–1010.
- Li, H.; Liefke, R.; Jiang, J.; Kurland, J.V.; Tian, W.; Deng, P.; Zhang, W.; He, Q.; Patel, D.J.; Bulyk, M.L.; et al. Polycomb-like proteins link the PRC2 complex to CpG islands. Nature 2017, 549, 287–291.
- Alekseyenko, A.A.; Gorchakov, A.A.; Kharchenko, P.V.; Kuroda, M.I. Reciprocal interactions of human C10orf12 and C17orf96 with PRC2 revealed by BioTAP-XL cross-linking and affinity purification. Proc. Natl. Acad. Sci. USA 2014, 111, 2488–2493.
- Conway, E.; Jerman, E.; Healy, E.; Ito, S.; Holoch, D.; Oliviero, G.; Deevy, O.; Glancy, E.; Fitzpatrick, D.J.; Mucha, M.; et al. A Family of Vertebrate-Specific Polycombs Encoded by the LCOR/LCORL Genes Balance PRC2 Subtype Activities. Mol. Cell 2018, 70, 408–421.
- Hauri, S.; Comoglio, F.; Seimiya, M.; Gerstung, M.; Glatter, T.; Hansen, K.; Aebersold, R.; Paro, R.; Gstaiger, M.; Beisel, C. A High-Density Map for Navigating the Human Polycomb Complexome. Cell Rep. 2016, 17, 583–595.
- Lee, C.H.; Holder, M.; Grau, D.; Saldaña-Meyer, R.; Yu, J.R.; Ganai, R.A.; Zhang, J.; Wang, M.; LeRoy, G.; Dobenecker, M.W.; et al. Distinct Stimulatory Mechanisms Regulate the Catalytic Activity of Polycomb Repressive Complex 2. Mol. Cell 2018, 70, 435–448.
- Kasinath, V.; Faini, M.; Poepsel, S.; Reif, D.; Feng, X.A.; Stjepanovic, G.; Aebersold, R.; Nogales, E. Structures of human PRC2 with its cofactors AEBP2 and JARID2. Science 2018, 359, 940–944.
- Sanulli, S.; Justin, N.; Teissandier, A.; Ancelin, K.; Portoso, M.; Caron, M.; Michaud, A.; Lombard, B.; da Rocha, S.T.; Offer, J.; et al. Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation. Mol. Cell 2015, 57, 769–783.
- Son, J.; Shen, S.S.; Margueron, R.; Reinberg, D. Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin. Genes Dev. 2013, 27, 2663–2677.
- Healy, E.; Mucha, M.; Glancy, E.; Fitzpatrick, D.J.; Conway, E.; Neikes, H.K.; Monger, C.; Van Mierlo, G.; Baltissen, M.P.; Koseki, Y.; et al. PRC2.1 and PRC2.2 Synergize to Coordinate H3K27 Trimethylation. Mol. Cell 2019, 76, 437–452.
- Perino, M.; van Mierlo, G.; Loh, C.; Wardle, S.M.T.; Zijlmans, D.W.; Marks, H.; Veenstra, G.J.C. Two Functional Axes of Feedback-Enforced PRC2 Recruitment in Mouse Embryonic Stem Cells. Stem Cell Reports 2020, 15, 1287–1300.
- Schwartz, Y.B.; Pirrotta, V. Polycomb silencing mechanisms and the management of genomic programmes. Nat. Rev. Genet. 2007, 8, 9–22.
- Grimaud, C.; Nègre, N.; Cavalli, G. From genetics to epigenetics: The tale of Polycomb group and trithorax group genes. Chromosome Res. 2006, 14, 363–375.
- Woo, C.J.; Kharchenko, P.V.; Daheron, L.; Park, P.J.; Kingston, R.E. A Region of the Human HOXD Cluster that Confers Polycomb-Group Responsiveness. Cell 2010, 140, 99–110.
- Simon, J.; Chiang, A.; Bender, W.; Shimell, M.J.; O’connor, M. Elements of the Drosophila Bithorax Complex That Mediate Repression by Polycomb Group Products. Dev. Biol. 1993, 158, 131–144.
- Van der Lugt, N.M.T.; Alkema, M.; Berns, A.; Deschamps, J. The Polycomb-group homolog Bmi-1 is a regulator of murine Hox gene expression. Mech. Dev. 1996, 58, 153–164.
- Pasini, D.; Bracken, A.P.; Hansen, J.B.; Capillo, M.; Helin, K. The Polycomb Group Protein Suz12 Is Required for Embryonic Stem Cell Differentiation. Mol. Cell. Biol. 2007, 27, 3769–3779.
- Faust, C.; Lawson, K.A.; Schork, N.J.; Thiel, B.; Magnuson, T. The Polycomb-group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development 1998, 125, 4495–4506.
- Kumar, B.; Navarro, C.; Winblad, N.; Schell, J.P.; Zhao, C.; Weltner, J.; Baqué-Vidal, L.; Salazar Mantero, A.; Petropoulos, S.; Lanner, F.; et al. Polycomb repressive complex 2 shields naïve human pluripotent cells from trophectoderm differentiation. Nat. Cell Biol. 2022, 24, 845–857.
- Zijlmans, D.W.; Talon, I.; Verhelst, S.; Bendall, A.; Van Nerum, K.; Javali, A.; Malcolm, A.A.; van Knippenberg, S.S.F.A.; Biggins, L.; To, S.K.; et al. Integrated multi-omics reveal polycomb repressive complex 2 restricts human trophoblast induction. Nat. Cell Biol. 2022, 24, 858–871.
- Del Mar Lorente, M.; Marcos-Gutiérrez, C.; Pérez, C.; Schoorlemmer, J.; Ramírez, A.; Magin, T.; Vidal, M. Loss- and gain-of-function mutations show a polycomb group function for Ring1A in mice. Development 2000, 127, 5093–5100.
- Voncken, J.W.; Roelen, B.A.; Roefs, M.; de Vries, S.; Verhoeven, E.; Marino, S.; Deschamps, J.; van Lohuizen, M. Rnf2 (Ring1b) deficiency causes gastrulation arrest and cell cycle inhibition. Proc. Natl. Acad. Sci. USA 2003, 100, 2468–2473.
- Takeuchi, T.; Yamazaki, Y.; Katoh-Fukui, Y.; Tsuchiya, R.; Kondo, S.; Motoyama, J.; Higashinakagawa, T. Gene trap capture of a novel mouse gene, jumonji, required for neural tube formation. Genes Dev. 1995, 9, 1211–1222.
- Højfeldt, J.W.; Hedehus, L.; Laugesen, A.; Tatar, T.; Wiehle, L.; Helin, K. Non-core Subunits of the PRC2 Complex Are Collectively Required for Its Target-Site Specificity. Mol. Cell 2019, 76, 423–436.
- Kloet, S.L.; Makowski, M.M.; Baymaz, H.I.; van Voorthuijsen, L.; Karemaker, I.D.; Santanach, A.; Jansen, P.W.T.C.; Di Croce, L.; Vermeulen, M. The dynamic interactome and genomic targets of Polycomb complexes during stem-cell differentiation. Nat. Struct. Mol. Biol. 2016, 23, 682–690.
- Beringer, M.; Pisano, P.; Di Carlo, V.; Blanco, E.; Chammas, P.; Vizán, P.; Gutiérrez, A.; Aranda, S.; Payer, B.; Wierer, M.; et al. EPOP Functionally Links Elongin and Polycomb in Pluripotent Stem Cells. Mol. Cell 2016, 64, 645–658.
