Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Sirius Huang and Version 1 by Olga A Koksharova.
Non-proteinogenic amino acid β-N-methylamino-L-alanine (BMAA) (syn: α-amino-β-methylaminopropionic acid, MeDAP; and 3-N-methyl-2,3-diaminopropanoic acid) was first isolated from the seeds of Cycas micronesica K.D.Hill (Cycadaceae). Non-proteinogenic amino acids (NPAAs) are not naturally encoded genetically and are not contained in the genetic code of any organism; however, they play diverse roles in prokaryotic and eukaryotic organisms. The research interest in BMAA arose due to the discovery of a link between chronic exposure to this diaminoacid and the occurrence of neurodegenerative diseases.
- algae
- cyanobacteria
- diatoms
- BMAA
1. History of BMAA Discovery
The prehistory that led to the discovery of the non-protein amino acid β-N-methylamino-L-alanine (BMAA) took place more than fifty years ago on Guam, an island in the western Pacific Ocean [2][1]. In the early 1950s, an extremely high level of neurodegenerative diseases was registered among the population of Guam; this indicator was 50–100 times higher than the global average [15][2]. To determine the cause of this anomaly, the researchers examined the brain tissue of Guam residents who died during this time period from the neurodegenerative disease Amyotrophic Lateral Sclerosis/Parkinsonism Dementia Complex (ALS-PDC) and found an interesting fact: brain tissue proteins were bound with a molecule that was identified as BMAA [7,8][3][4]. The search for the source of BMAA had been started.
However, only ten years later, in 1967, BMAA was finally (for the first time) isolated from the seeds of queen sago (Cycas circinalis) [5[5][6],6], a plant whose seeds had been used for decades in the local diet of the inhabitants of Guam. Later, in the early 2000s, BMAA was discovered in colonies of symbiotic cyanobacterial species—Nostoc and Anabaena, which were localized within the coralloid roots of C. circinalis, and therefore it was suggested that these symbiotic cyanobacteria are the original producers of BMAA and an indirect cause of profound human neurodegenerative disease (which is now known to be caused by the long-term bioaccumulation of BMAA in brain tissue) [9,10,11,12,16][7][8][9][10][11].
2. BMAA Producers
2.1. Cyanobacteria
BMAA can be synthesized by both free-living and symbiotic species of cyanobacteria [17][13]. Moreover, BMAA has been found not only in filamentous cyanobacterial species but also in various unicellular cyanobacteria. Cyanobacteria producing BMAA are widespread. They can be found in various types of habitats—marine, freshwater, and terrestrial [18,19,20,21,22,23,24,25,26,27][14][15][16][17][18][19][20][21][22][23]. Strains of cyanobacteria producing BMAA have been collected all over the world—in South Africa [18][14], Hawaii [19][15], India, and Australia [17][13], Peru [20][16], Great Britain [21][17], the United States [22][18], the Swedish waters of Baltic Sea [23][19], Portugal [24][20], and Germany [25][21].
Unexpectedly, it turned out that isomeric forms of BMAA can also be found in cyanobacteria, such as 2,4-diaminobutyric acid (2,4-DAB) and N-(2-aminoethyl) glycine (AEG). These isomeric amino acids were found in various species of freshwater cyanobacteria: Anabaena, Leptolyngbya sp., Oscillatoria sp., Merismopedia sp., and Microcystis aeruginosa [28][24]. In addition, BMAA and its isomers have been identified even in 100-year-old Antarctic cyanobacterial mats that are still preserved in the herbarium of the Natural History Museum in London, UK [29][25].
2.2. Diatoms
While many studies have shown that BMAA is produced by cyanobacteria, only a few studies have addressed this issue in relation to eukaryotic groups of algae. Several studies have shown that BMAA-production occurs in marine and fresh-water diatoms [30,31,32,33][26][27][28][29].
The first proof of the fact that BMAA is not only produced by prokaryotic cyanobacteria but can also be synthesized by eukaryotic organisms, such as diatoms, was provided in 2014 by L. Jiang and colleagues in the laboratory of Ulla Rasmussen [30][26]. In the mentioned study, BMAA was detected in nanogram amounts (1.0–3.8 ng/g dry weight) in six different axenic diatom cultures, viz. Navicula pelliculosa (CCAP 1050/9), Thalassiosira sp. (CCAP 1085/15), Achnanthes sp. (CCAP 1095/1), Skeletonema marinoi SAAE08603, Skeletonema marinoi ST28, and Proboscia inermis (CCAP 1064/1). In addition, BMAA was also found in field samples of plankton that were collected on the west coast of the Swedish Baltic Sea and in which marine diatoms were found in overly representative amounts.
In the following years, several more studies showed that both marine and freshwater diatoms can produce BMAA [31,32,33][27][28][29]. Four marine diatoms, Phaeodactylum tricornutum, Chaetoceros sp., Chaetoceros calcitrans and Thalassiosira pseudonana were studied [31][27], and for all four species of diatoms, a direct dependence of the concentration of the total amount of soluble BMAA and DAB on the growth phase rate was demonstrated. The highest concentration of BMAA was found during the stationary growth phase of all these studied marine species. This finding allowed the authors to assume that BMAA is a secondary metabolite of diatoms. Later, in another work [32][28], it was revealed that freshwater diatoms can also produce BMAA, as well as its isomers AEG and 2,4-DAB.
2.3. Dinoflagellates
Dinoflagellates are known as a widespread group of plankton living in both marine and fresh waters. Thus far, only one paper [34][30] has reported that a laboratory-grown culture of dinoflagellate Gymnodinium catenatum is capable of producing BMAA. It should be noted that BMAA was found in dinoflagellates in a considerably high level (0.457 ± 0.186 µg/g DW). More research is needed to confirm this discovery.
2.4. Bacteria
In 1975, during the search for new antibiotics, BMAA was discovered as a molecular component of peptides that were found to be produced by Paenibacillus pulvifaciens, a widespread facultative anaerobic endospore-forming bacterium [14][31]. Then, in 2014, peptides of a similar structure (viz. with a BMAA in the structure) were isolated from Paenibacillus larvae, another species within the Paenibacillus genius [14][31].
Paenibacillus spp. is a widespread group of bacteria (more than 200 species of Paenibacillus spp. are known), which is the cause of a highly infectious disease of bees, called American foulbrood. The fact that BMAA was found in the structure of peptides synthesized by Paenibacillus spp. should be taken into account, considering the common distribution of these bacteria and the fact that some species of Paenibacillus spp. have been detected even in food groceries and human interstitial fluids [14][31]; therefore, the presence of peptide-associated BMAA may pose a hidden threat to consumer (in case BMAA is released) and should be thoroughly studied in the future.
2.5. Plants
Can higher plants synthesize BMAA? The knowledge on this issue is still limited. In only one study, BMAA was found in various tissues of plant Cycas micronesica (a plant that is known to have the ability to form a symbiosis with cyanobacteria); however, at the time of the study (according to the authors) did not have symbiotic strains of cyanobacteria on the roots [35][32]. It was stated that C. micronesica seedlings were grown without endophytic cyanobacterial symbiosis. During the growth period, BMAA was quantified at different time points in various parts of the plant. The amount of BMAA increased by 79% during the nine months of seedling growth, and the root tissue contained 75% of the total amount of BMAA [35][32].
However, a number of questions arise in connection with these findings. How is BMAA synthesis related to plant age and to (laboratory or natural) growth conditions? Are only Cycas micronesica able to synthesize this amino acid? Or can other higher plants also do this? Without a doubt, it is necessary to conduct additional research on this issue in order to find an answer to the question: can plants really synthesize and accumulate BMAA in the absence of cyanobacteria?
Summing up all the above, it can be stated that the current experimental data have shown that various photoautotrophic organisms can produce BMAA, as well as some strains of bacteria. The ability of various species of cyanobacteria and microalgae to synthesize BMAA in a wide range of concentrations—from nanograms to thousands of micrograms per gram of dry weight [17,30,31][13][26][27]—may indicate the biological significance of the role that this molecule plays in the life of these species [36,37,38][33][34][35]. What kind of role could it be? More research is needed to find the answer and confirm it.
3. Bioaccumulation and Biomagnification
In the last decade, researchers have demonstrated that BMAA, which is synthesized by phytoplankton in aquatic ecosystems, is further transmitted along the food chain to zooplankton and other invertebrates (mussels, oysters, and shrimps) and eventually accumulates in the brain and muscle tissue of fish, as well as in the brains of dolphins (see Section 4.2) and chicken tissues [14,23,39,40,41,42,43,44,45][19][31][36][37][38][39][40][41][42] (Table 1).
Table 1.
BMAA has been detected in a range of organisms (some examples are presented).
| Source | Concentration of BMAA μg/g Dry Weight of Sample |
Method of Identification | Reference |
|---|---|---|---|
| Cyanobacteria Nostoc | 0.3 | HPLC-MS | [9][7] |
| Cycad/cyanobacterial symbiosis | 37–1161 | HPLC-MS | [9][7] |
| the immature male sporangi of Cycas micronesica |
1546 | HPLC-MS | [9][7] |
| the outer integument layer of the cycas seed sarcotesta | 1161 | HPLC-MS | [9][7] |
| Flying Foxes | 3556 | HPLC-MS | [9][7] |
| brain tissues from the frontal cortex of six Chamorro patients | 6 | HPLC-MS | [9][7] |
| brain tissues from the frontal cortex of two Alzheimer’s patients from Canada | 6.6 | HPLC-MS | [9][7] |
| Azolla filiculoides with cyanobacterial symbionts in its leaves |
2 | HPLC-MS | [9][7] |
| Gunnera kauaiensi with cyanobacterial symbionts in its leaves |
4 | HPLC-MS | [9][7] |
| Nostoc PCC 9305 (symbiont of Anthoceros ) |
156 (free) 1400 (protein-bound) |
HPLC-MS | [17][13] |
| Nostoc PCC 7422 (symbiont of Cycas) |
962 (protein-bound) | HPLC-MS | [17][13] |
| Nostoc 8001 (symbiont of Gunnera monoica) |
203 (free) 664 (protein-bound) |
HPLC-MS | [17][13] |
| Microcystis PCC 7820 (Freshwater, Scotland) |
6 (free) 12 (protein-bound) |
HPLC-MS | [17][13] |
| Chroococcidiopsis indica GQ2-7 (Marine coral) | 435 (free) 76 (protein-bound) |
HPLC-MS | [17][13] |
| Chroococcidiopsis indica GT-3-26 (Marine rock) |
1306 (free) 5415 (protein-bound) |
HPLC-MS | [17][13] |
| Cylindrospermopsis raciborskii CR3 (Freshwater, Australia) |
6478 (free) 14 (protein-bound) |
HPLC-MS | [17][13] |
| Nostoc 268 (Brackish Water, Baltic Sea) |
34 (free) 274 (protein-bound) |
HPLC-MS | [17][13] |
| Nostoc sp. CMMED 01 (Marine, Hawaiian Islands) |
1243 (free) 1070 (protein-bound) |
HPLC-MS | [17][13] |
| Chlorogloeopsis PCC 6912 (Soil, India) |
758 (free) | HPLC-MS | [17][13] |
| Fischerella PCC 7521 (Yellowstone, hot spring, USA) |
44 (free) 175 (protein-bound) |
HPLC-MS | [17][13] |
| Scytonema PCC 7110 (Limestone cave, Bermuda) |
1733 (protein-bound) | HPLC-MS | [17][13] |
| Mytilus edulis (common mussel, Sweden’s west coast) |
0.15–0.2 | HPLC-MS/MS | [23][19] |
| Ostrea edulis (common oyster, Sweden’s west coast ) | 0.006–0.14 | HPLC-MS/MS | [23][19] |
| Scophthalmus maximus (turbot) (Brain tissue) |
0.047–1.29 | HPLC-MS/MS | [23][19] |
| Microcystis PCC7806 and Synechocystis J341 |
the relative abundance of labeled amino acids based on LC/MS/MS peak areas | stable isotope 15N and UPLC-MS/MS | [27][23] |
| Thalassiosira sp. (CCAP 1085/15) diatom |
0.0033 | UHPLC-MS/MS | [30][26] |
| Skeletonema marinoi isolates (SAAE 08603) diatom |
0.0011 | UHPLC-MS/MS | [30][26] |
| Phaeodactylum tricornutum diatom |
0.20–1.4 | HPLC-MS/MS | [31][27] |
| Cyclotella (diatom from Lake Liddell ) |
0.103 (free) 0.154 (protein-bound) |
LC-MS/MS | [32][28] |
| Navicula (from Lostock Dam) |
0.151 (free) 0.369 (protein-bound) |
LC-MS/MS | [32][28] |
| Cycas micronesica K.D. Hill Gametophyte Southern |
1.94 | GC-MS | [35][32] |
| Cycas micronesica K.D. Hill Gametophyte Nothern |
3.32 | GC-MS | [35][32] |
| Cycas micronesica K.D. Hill total plant Southern |
3.44 | GC-MS | [35][32] |
| Cycas micronesica K.D. Hill total plant Nothern |
4.9 | GC-MS | [35][32] |
| Blue mussel (Mytilus edulis) Sweden (west coast) |
0.08–0.9 | UHPLC-MS/MS | [39][36] |
| Oyster (Ostrea edulis) Greece |
0.32 | UHPLC-MS/MS | [39][36] |
| Oyster (Crassostrea gigas) France |
0.66 | UHPLC-MS/MS | [39][36] |
| Triticum aestivum (mature seeds) via irrigation |
217 ± 150 ng g FW−1 (protein-bound) |
UHPLC-MS/MS | [43][40] |
| chicken tissues (i.e., muscle, liver, brain, and eye) |
0.0045–0.03 (free) 0.03–0.23 (protein-bound) |
UHPLC-MS/MS | [45][42] |
The transfer of BMAA from an aquatic ecosystem to a terrestrial ecosystem can occur, as was demonstrated in [43[40][41][42],44,45], and is caused by irrigation of fields by spraying, in which BMAA accumulates in plant seeds and plant tissues. Finally, passing through various food webs, this amino acid eventually reaches higher animals, including humans [10,11,22,23,40,45][8][9][18][19][37][42] (Figure 1). Protein-associated BMAA molecules were found in significant concentrations (31–356 µg/g, [16][11]) in the brain tissues of patients who died from ALS-PDC and Alzheimer’s disease (AD) [11,16][9][11].
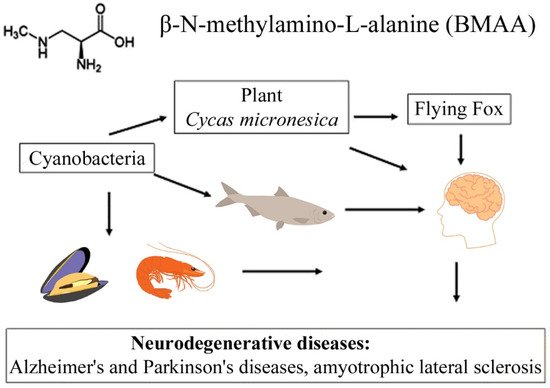
Figure 1.
This diagram shows the bioaccumulation pathways of BMAA.
A convincing example is the bioaccumulation of BMAA in the food chains of the Guam ecosystem [10,12][8][10]. It was found that, in this food chain, concentrations of free and protein-bound BMAA increased drastically during the transition from cyanobacteria (0.3 µg/g and 72 µg/g) to cycad seeds (9 µg/g and 89 µg/g), and then to the Micronesian flying fox Pteropus mariannus (3556 µg/g and 146 µg/g) [10][8]; and an even higher concentration of protein-bound BMAA (627 µg/g) was detected in the postmortem brain tissue of ALS patients as a result of biomagnification [10][8].
It is clear that the facts regarding the bioaccumulation of BMAA are alarming and further research should be conducted to study the mechanisms of synthesis of this molecule, to detect the conditions in which this synthesis occurs, and to clarify the biological role of this compound in the life of organisms producing BMAA.
4. Biosynthesis Pathways of BMAA: Different Hypothesis
There are several hypotheses about the possible ways of BMAA biosynthesis [14,38,46,47][31][35][43][44]. BMAA can occur in bacterial cells as a result of cellular metabolic processes [14,38][31][35]. Nunn and Codd discussed possible metabolic pathways involved in the biosynthesis of BMAA in cyanobacteria [38][35]. The authors suggested that BMAA can be formed by methylation of 2,3-diaminopropanoic acid (2,3-DAP) within a macromolecular structure of the multi-enzyme complexes and subsequently released in free form. The first hypothesis was put forward in 2003 by Brenner et al., who suggested a possible pathway for the synthesis of BMAA in the Cycas rumphii plant, which was supported by the analysis of expressed sequence tags (EST) from C. rumphii [46][43].
The authors suggested that, due to the structural similarity of BMAA with other beta-substituted alanines, such as phosphoserine, cysteine, o-acetylserine, or β-Cyano-L-alanine, alanine can be used for the biosynthesis of BMAA in a two-stage reaction. The first stage of the reaction is the transfer of NH3 to the β-carbon site of alanine that is preformed by a cysteine synthase-like protein. The authors identified two candidate genes, which encode cysteine synthase (Gene Bank accession numbers CB089577 and CB092214). The second stage of the reaction consists of the methylation of the beta-amino group of beta-substituted alanine.
The EST library contains two genes encoding potential methyltransferases: caffeic acid O-methyltransferase II and caffeoyl-CoA 3-O-methyltransferase, which are encoded by CB091906 and CB090738, respectively. These genes are necessary in order to catalyze the second stage of BMAA biosynthesis [46][43]. Later, in the first review [13][12], which was devoted to discussing the biological roles of BMAA, it was suggested that the search for homologous genes in the genomes of cyanobacteria and diatoms is of interest, since it can contribute to elucidating the mechanisms of BMAA synthesis in these organisms.
Most recently, the first such genomic study was conducted [47][44]. In this interesting study, the authors used bioinformatics tools to explore hypotheses regarding BMAA biosynthesis in cyanobacteria by assessing the presence or absence of enzymes in six known potential metabolic pathways in 130 cyanobacterial genomes. It has been shown that most of the enzymes involved in the pathways leading to the putative precursor of BMAA (2,3-diaminopropanoic acid, 2,3-DAP) in other species have not been detected in cyanobacteria. Only genes encoding SbnA and SbnB were found in a limited subset of cyanobacterial species.
Due to the coordinated action of these proteins, the biosynthesis of 2,3-DAP occurs in Gram-positive bacteria Staphylococcus aureus Rosenbach 1884. The authors emphasized the potential physiological role of 2,3-DAP in the formation of siderophores in some cyanobacteria species and showed that the pam gene cluster responsible for directing the biosynthesis of the peptide-bound BMAA in Paenibacillus larvae (Paenibacillaceae), was not found in the genomes of 130 species of cyanobacteria and was also not found in 93 genomes of Paenibacillus (except of P. larvae).
This study also showed that the presence in some cyanobacteria species of genes presumably encoding the enzymes 2,3-diaminopropionate ammonia lyase (DAPAL, EC 4.3.1.15) and reactive intermediate deaminase A (RIDA, EC 3.5.99.10) may explain the inability to detect 2,3-DAP in analytical studies. DAPAL is a prokaryotic type II PLP-dependent enzyme that catalyses the degradation of R- and S-forms of 2,3-DAP to 2-aminoacrylate and ammonium.
The biosynthesis of 2,3-DAP in cyanobacteria seems to be either limited to a small subset of cyanobacterial species, or there may be many additional ways of biosynthesis of this amino acid. The authors believe that it is also possible that cyanobacteria synthesize BMAA using a pathway or pathways that have not yet been discovered.
Another possible source of BMAA was recently discussed in [14][31]. Bacteria Paenibacillus pulvifaciens and Paenibacillus larvae contain antimicrobial peptides (galantins and paenilamicins), which include BMAA (Figure 2). In cells of P. larvae, BMAA occurs within an NRPS-multienzyme complex by the methylation of the 2,3-DAP residue. A cluster of pam genes necessary for the biosynthesis of paenilamicins was identified [48][45]. Nunn and Codd suggested that free BMAA does not occur in the cells of P. larvae. This bacterium is responsible for a highly infectious disease of bees.
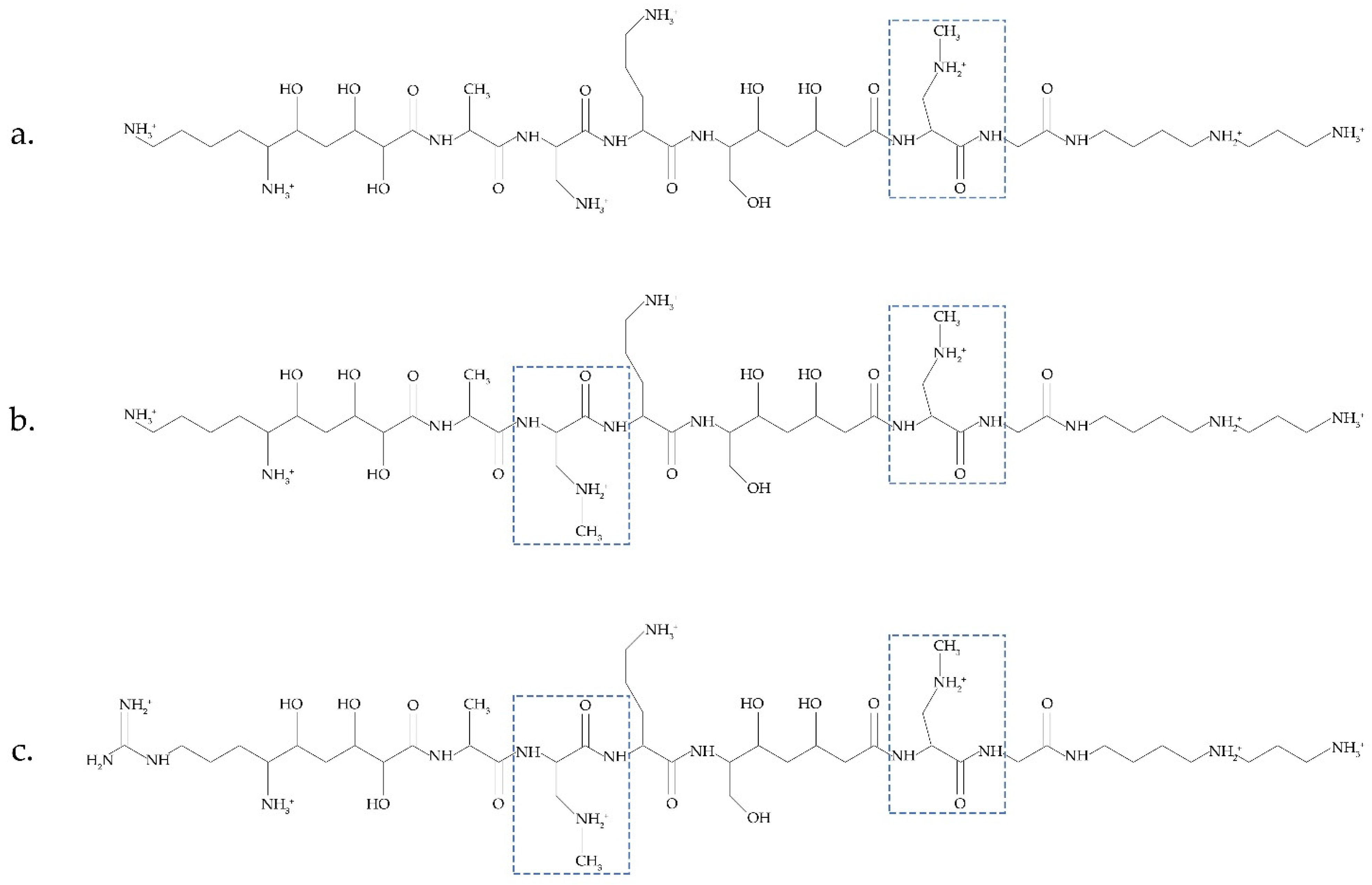
Figure 2.
Antimicrobial peptides (galantin (
a
) and paenilamicins (
b
,
A metabolic turnover of paenilamicin antibiotics and their hydrolysis by peptidases can occur in the intestines of honeybee larvae. As a result, a free BMAA is released. Considering that Paenibacillus species are always present in the environment and are used agriculturally and that some Paenibacillus species infect human food and have even been isolated from human body fluids [14][31], it can be concluded that it is necessary to investigate these bacteria and their peptides in detail as another important source of BMAA. Considering that cyanobacteria also synthesize biologically active toxic peptides [49][46], it will be interesting to clarify whether cyanobacteria can also have BMAA as a peptide component.
It is important to keep in mind the possibility of spore formation by many bacteria, including Paenibacillus spp. There is a medical hypothesis (the so-called “spore hypothesis”) that explains both early-onset and late-onset Parkinson’s disease [50][47]. There is a hypothetical probability of the formation of BMAA-endospores from Paenibacillus spp. that can penetrate into the central nervous system [14][31]. In addition, we should consider the possibility of the existence of other BMAA-producing bacteria that are part of the human microbiome [14][31].
The production of BMAA by representatives of the intestinal microbiota has been hypothesized as another possible pathway of chronic exposure to BMAA [51,52][48][49]. This assumption should be investigated. One of the potential candidates for the role of such an “internal enemy” is Melainabacteria, a non-photosynthetic phylogenetic clade of divergent cyanobacteria identified as representatives of the human gut microbiota [53,54][50][51]. These hypotheses under discussion are waiting for their research and confirmation or refutation.
References
- Nunn, P.B. 50 years of research on α-amino-β-methylaminopropionic acid (β-methylaminoalanine). Phytochemistry 2017, 144, 271–281.
- Brody, J.A.; Stanhope, J.M.; Kurland, L.T. Patterns of amyotrophic lateral sclerosis and parkinsonism-dementia on Guam. Contemp Neurol Ser. 1975, 12, 45–70.
- Hirano, A.; Malamud, N.; Elizan, T.S.; Kurland, L.T. Amyotrophic lateral sclerosis and Parkinsonism-dementia complex on Guam. Further pathologic studies. Arch. Neurol. 1966, 15, 35–51.
- Spencer, P.S.; Nunn, P.B.; Hugon, J.; Ludolph, A.C.; Ross, S.M.; Roy, D.N.; Robertson, R.C. Guam amyotrophic lateral sclerosis-parkinsonism-dementia linked to a plant excitant neurotoxin. Science 1987, 237, 517–522.
- Casagrande, D.J.; Given, P.H. Geochemistry of amino acids in some Florida peat accumulation-II. Amino acid distributions. Geochim. Cosmochim. Acta 1980, 44, 1493–1507.
- Walsh, C.T.; O’Brien, R.V.; Khosla, C. Nonproteinogenic Amino Acid Building Blocks for Nonribosomal Peptide and Hybrid Polyketide Scaffolds. Angew. Chem. Int. Ed. 2013, 52, 7098–7124.
- Cox, P.A.; Banack, S.A.; Murch, S.J. Biomagnification of cyanobacterial neurotoxins and neurodegenerative disease among the Chamorro people of Guam. Proc. Natl. Acad. Sci. USA 2003, 100, 13380–13383.
- Murch, S.J.; Cox, P.A.; Banack, S.A. A mechanism for slow release of biomagnified cyanobacterial neurotoxins and neurodegenerative disease in Guam. Proc. Natl. Acad. Sci. USA 2004, 101, 12228–12231.
- Murch, S.J.; Cox, P.A.; Banack, S.A.; Steele, J.C.; Sacks, O.W. Occurrence of beta-methylamino-l-alanine (BMAA) in ALS/PDC patients from Guam. Actaneurologica Scand. 2004, 110, 267–269.
- Bradley, W.G.; Mash, D.C. Beyond Guam: The cyanobacteria/BMAA hypothesis of the cause of ALS and other neurodegenerative diseases. Amyotroph. Lateral Scler. 2009, 10, 7–20.
- Pablo, J.; Banack, S.A.; Cox, P.A.; Johnson, T.E.; Papapetropoulos, S.; Bradley, W.G.; Buck, A.; Mash, D.C. Cyanobacterial neurotoxin BMAA in ALS and Alzheimer’s disease. Acta Neurol Scand. 2009, 120, 216–225.
- Popova, A.A.; Koksharova, O.A. Neurotoxic non-proteinogenic amino acid β-N-methylamino-L-alanine and its role in biological systems. Biochemistry 2016, 81, 794–805.
- Cox, P.A.; Banack, S.A.; Murch, S.J.; Rasmussen, U.; Tien, G.; Bidigare, R.R.; Metcalf, J.S.; Morrison, L.F.; Codd, G.A.; Bergman, B. Diverse taxa of cyanobacteria produce beta-N-methylamino-L-alanine, a neurotoxic amino acid. Proc. Natl. Acad. Sci. USA 2005, 102, 5074–5078.
- Esterhuizen, M.; Downing, T.G. β-N-methylamino-L-alanine (BMAA) in novel South African cyanobacterial isolates. Ecotoxicol. Environ. Saf. 2008, 71, 309–313.
- Banack, S.A.; Johnson, H.E.; Cheng, R.; Cox, P.A. Production of the neurotoxin BMAA by a marine cyanobacterium. Mar. Drugs 2007, 5, 180–196.
- Johnson, H.E.; King, S.R.; Banack, S.A.; Webster, C.; Callanaupa, W.J.; Cox, P.A. Cyanobacteria (Nostoc commune) used as a dietary item in the Peruvian highlands produce the neurotoxic amino acid BMAA. J. Ethnopharmacol. 2008, 118, 159–165.
- Metcalf, J.S.; Banack, S.A.; Lindsay, J.; Morrison, L.F.; Cox, P.A.; Codd, G.A. Co-occurrence of beta-N-methylamino-L-alanine, a neurotoxic amino acid with other cyanobacterial toxins in British waterbodies, 1990–2004. Environ. Microbiol. 2008, 10, 702–708.
- Brand, L.E.; Pablo, J.; Compton, A.; Hammerschlag, N.; Mash, D.C. Cyanobacterial blooms and the occurrence of the neurotoxin beta-N-methylamino-L-alanine (BMAA) in South Florida aquatic food webs. Harmful Algae 2010, 9, 620–635.
- Jonasson, S.; Eriksson, J.; Berntzon, L.; Spacil, Z.; Ilag, L.L.; Ronnevi, L.O.; Rasmussen, U.; Bergman, B. Transfer of a cyanobacterial neurotoxin within a temperate aquatic ecosystem suggests pathways for human exposure. Proc. Natl. Acad. Sci. USA 2010, 107, 9252–9257.
- Cervantes Cianca, R.C.; Baptista, M.S.; Lopes, V.R.; Vasconcelos, V.M. The non-protein amino acid β-N-methylamino-L-alanine in Portuguese cyanobacterial isolates. Amino Acids 2012, 42, 2473–2479.
- Contardo-Jara, V.; Sebastian Funke, M.; Peuthert, A.; Pflugmacher, S. β-N-Methylamino-L-alanine exposure alters defense against oxidative stress in aquatic plants Lomariopsis lineata, Fontinalis antipyretica, Riccia fluitans and Taxiphyllum barbieri. Ecotoxicol. Environ. Saf. 2013, 88, 72–78.
- Spacil, Z.; Eriksson, J.; Jonasson, S.; Rasmussen, U.; Ilag, L.L.; Bergman, B. Analytical protocol for identification of BMAA and DAB in biological samples. Analyst 2009, 135, 127–132.
- Downing, S.; Banack, S.A.; Metcalf, J.S.; Cox, P.A.; Downing, T.G. Nitrogen starvation of cyanobacteria results in the production of β-N-methylamino-L-alanine. Toxicon 2011, 58, 187–194.
- Violi, J.P.; Mitrovic, S.M.; Colville, A.; Main, B.J.; Rodgers, K.J. Prevalence of β-methylamino-L-alanine (BMAA) and its isomers in freshwater cyanobacteria isolated from eastern Australia. Ecotoxicol Environ Saf. 2019, 172, 72–81.
- Jungblut, A.D.; Wilbraham, J.; Banack, S.A.; Metcalf, J.S.; Codd, G.A. Microcystins, BMAA and BMAA isomers in 100-year-old Antarctic cyanobacterial mats collected during Captain R.F. Scott’s Discovery Expedition. Eur. J. Phycol. 2018, 53, 115–121.
- Jiang, L.; Eriksson, J.; Lage, S.; Jonasson, S.; Shams, S.; Mehine, M.; Ilag, L.L.; Rasmussen, U. Diatoms: A novel source for the neurotoxin BMAA in aquatic environments. PLoS ONE 2014, 9, e84578.
- Réveillon, D.; Séchet, V.; Hess, P.; Amzil, Z. Production of BMAA and DAB by diatoms (Phaeodactylum tricornutum, Chaetoceros sp., Chaetoceros calcitrans and, Thalassiosira pseudonana) and bacteria isolated from a diatom culture. Harmful Algae 2016, 58, 45–50.
- Violi, J.P.; Facey, J.A.; Mitrovic, S.M.; Colville, A.; Rodgers, K.J. Production of β-methylamino-L-alanine (BMAA) and Its Isomers by Freshwater Diatoms. Toxins 2019, 11, 512.
- Bates, S.S.; Lundholm, N.; Hubbard, K.A.; Montresor, M.; Leaw, C.P. Toxic and harmful marine diatoms. In Diatoms: Fundamentals and Applications; Seckbach, J., Gordon, R., Eds.; Scrivener Publishing LLC: Salem, MA, USA, 2019; pp. 389–434.
- Lage, S.; Costa, P.R.; Moita, T.; Eriksson, J.; Rasmussen, U.; Rydberg, S.J. BMAA in shellfish from two Portuguese transitional water bodies suggests the marine dinoflagellate Gymnodinium catenatum as a potential BMAA source. Aquat. Toxicol. 2014, 152, 131–138.
- Nunn, P.B.; Codd, G.A. Environmental distribution of the neurotoxin l-BMAA in Paenibacillus species. Toxicol. Res. 2019, 8, 781–783.
- Marler, T.E.; Snyder, L.R.; Shaw, C.A. Cycas micronesica (Cycadales) plants devoid of endophytic cyanobacteria increase in β-methylamino-L-alanine. Toxicon 2010, 56, 563–568.
- Downing, T.G.; Phelan, R.R.; Downing, S. A potential physiological role for cyanotoxins in cyanobacteria of arid environments. J. Arid Environ. 2015, 112, 147–151.
- Downing, S.; Downing, T.G. The metabolism of the non proteinogenic amino acid β-N-methylamino-L-alanine (BMAA) in the cyanobacterium Synechocystis PCC 6803. Toxicon 2016, 115, 41–48.
- Nunn, P.B.; Codd, G.A. Metabolic solutions to the biosynthesis of some diaminomonocarboxylic acids in nature: Formation in cyanobacteria of the neurotoxins 3-N-methyl-2,3-diaminopropanoic acid (BMAA) and 2,4-diaminobutanoic acid (2,4-DAB). Phytochemistry 2017, 144, 253–270.
- Jiang, L.; Kiselova, N.; Rosén, J.; Ilag, L.L. Quantification of neurotoxin BMAA (β-N-methylamino-L-alanine) in seafood from Swedish markets. Sci. Rep. 2014, 4, 6931.
- Salomonsson, M.L.; Fredriksson, E.; Alfjorden, A.; Hedeland, M.; Bondesson, U. Seafood sold in Sweden contains BMAA: A study of free and total concentrations with UHPLC-MS/MS and dansyl chloride derivatization. Toxicol. Rep. 2015, 2, 1473–1481.
- Hammerschlag, N.; Davis, D.A.; Mondo, K.; Seely, M.S.; Murch, S.J.; Glover, W.B.; Divoll, T.; Evers, D.C.; Mash, D.C. Cyanobacterial Neurotoxin BMAA and Mercury in Sharks. Toxins 2016, 8, 238.
- Regueiro, J.; Negreira, N.; Carreira-Casais, A.; Pérez-Lamela, C.; Simal-Gándara, J. Dietary exposure and neurotoxicity of the environmental free and bound toxin β-N-methylamino-l-alanine. Food Res. Int. 2017, 100, 1–13.
- Contardo-Jara, V.; Schwanemann, T.; Esterhuizen-Londt, M.; Pflugmacher, S. Protein association of β-N-methylamino-L-alanine in Triticum aestivum via irrigation. Food Addit. Contam. Part A 2018, 35, 731–739.
- Esterhuizen-Londt, M.; Pflugmacher, S. Vegetables cultivated with exposure to pure and naturally occurring β-N-methylamino-L-alanine (BMAA) via irrigation. Environ. Res. 2019, 169, 357–361.
- Kim, S.-Y.; Rydberg, S. Transfer of the Neurotoxin β-N-methylamino-l-alanine (BMAA) in the Agro–Aqua Cycle. Mar. Drugs 2020, 18, 244.
- Brenner, E.D.; Stevenson, D.W.; McCombie, R.W.; Katari, M.; A Rudd, S.; Mayer, K.F.X.; Palenchar, P.M.; Runko, S.J.; Twigg, R.W.; Dai, G.; et al. Expressed sequence tag analysis in Cycas, the most primitive living seed plant. Genome Biol. 2003, 4, R78.
- Mantas, M.J.Q.; Nunn, P.B.; Codd, G.A.; Barker, D. Genomic insights into the biosynthesis and physiology of the cyanobacterial neurotoxin 3-N-methyl-2,3-diaminopropanoic acid (BMAA). Phytochemistry 2022, 200, 113198.
- Müller, S.; Garcia-Gonzalez, E.; Mainz, A.; Hertlein, G.; Heid, N.C.; Mösker, E.; van den Elst, H.V.; Overkleeft, H.S.; Genersch, E.; Süssmuth, R.D.; et al. Paenilamicin: Structure and biosynthesis of a hybrid nonribosomal peptide/polyketide antibiotic from the bee pathogen Paenibacillus larvae. Angew. Chem. Int. Ed. 2014, 53, 10821–10825.
- Du, X.; Liu, H.; Yuan, L.; Wang, Y.; Ma, Y.; Wang, R.; Chen, X.; Losiewicz, M.; Guo, H.; Zhang, H. The Diversity of Cyanobacterial Toxins on Structural Characterization, Distribution and Identification: A Systematic Review. Toxins 2019, 11, 530.
- Berstad, K.; Berstad, J.E.R. Parkinson’s disease; the hibernating spore hypothesis. Med. Hypotheses 2017, 104, 48–53.
- Brenner, S. Blue-green algae or cyanobacteria in the intestinal micro-flora may produce neurotoxins such as β-N-Methylamino-L-Alanine (BMAA) which may be related to development of amyotrophic lateral sclerosis, Alzheimer’s disease and Parkinson-Dementia-Complex in humans and equine motor neuron disease in horses. Med. Hypotheses 2013, 80, 103.
- Nunes-Costa, D.; Magalhães, J.D.; G-Fernandes, M.; Cardoso, S.M.; Empadinhas, N. Microbial BMAA and the Pathway for Parkinson’s Disease Neurodegeneration. Front. Aging Neurosci. 2020, 12, 26.
- Di Rienzi, S.C.; Sharon, I.; Wrighton, K.C.; Koren, O.; Hug, L.A.; Thomas, B.C.; Goodrich, J.K.; Bell, J.; Spector, T.D.; Banfield, J.T.; et al. The human gut and groundwater harbor non-photosynthetic bacteria belonging to a new candidate phylum sibling to Cyanobacteria. Elife 2013, 2, e01102.
- Soo, R.M.; Skennerton, C.T.; Sekiguchi, Y.; Imelfort, M.; Paech, S.J.; Dennis, P.G.; Steen, J.A.; Parks, D.H.; Tyson, G.W.; Hugenholtz, P. An expanded genomic representation of the phylum cyanobacteria. Genome. Biol. Evol. 2014, 6, 1031–1045.
More
