Antimicrobial resistance (AMR) is today universally recognised as a global threat because of the rapid emergence and dissemination of resistant bacteria and genes among humans, animals, and the environment on a global scale. AMR thus represents a heavy burden for healthcare systems all over the world. ESKAPE pathogens (Enterococcus faecium, Staphylococcus aureus, Klebsiella pneumoniae, Acinetobacter baumannii, Pseudomonas aeruginosa et Enterobacter spp.) combined with antibiotic resistance have greatly increased the risk of morbidity and mortality, especially in ICU settings. Lateral flow assays (LFA) are inexpensive, rapid, and efficient tools that are easy to implement in the routine workflow of laboratories as new first-line tests against AMR with bacterial colonies, and in the near future directly with biological media.
- LFA
- detection
- antibiotic resistance
1. General Presentation of Lateral FIlow ImmunoAssay
1.1. Components and Principle
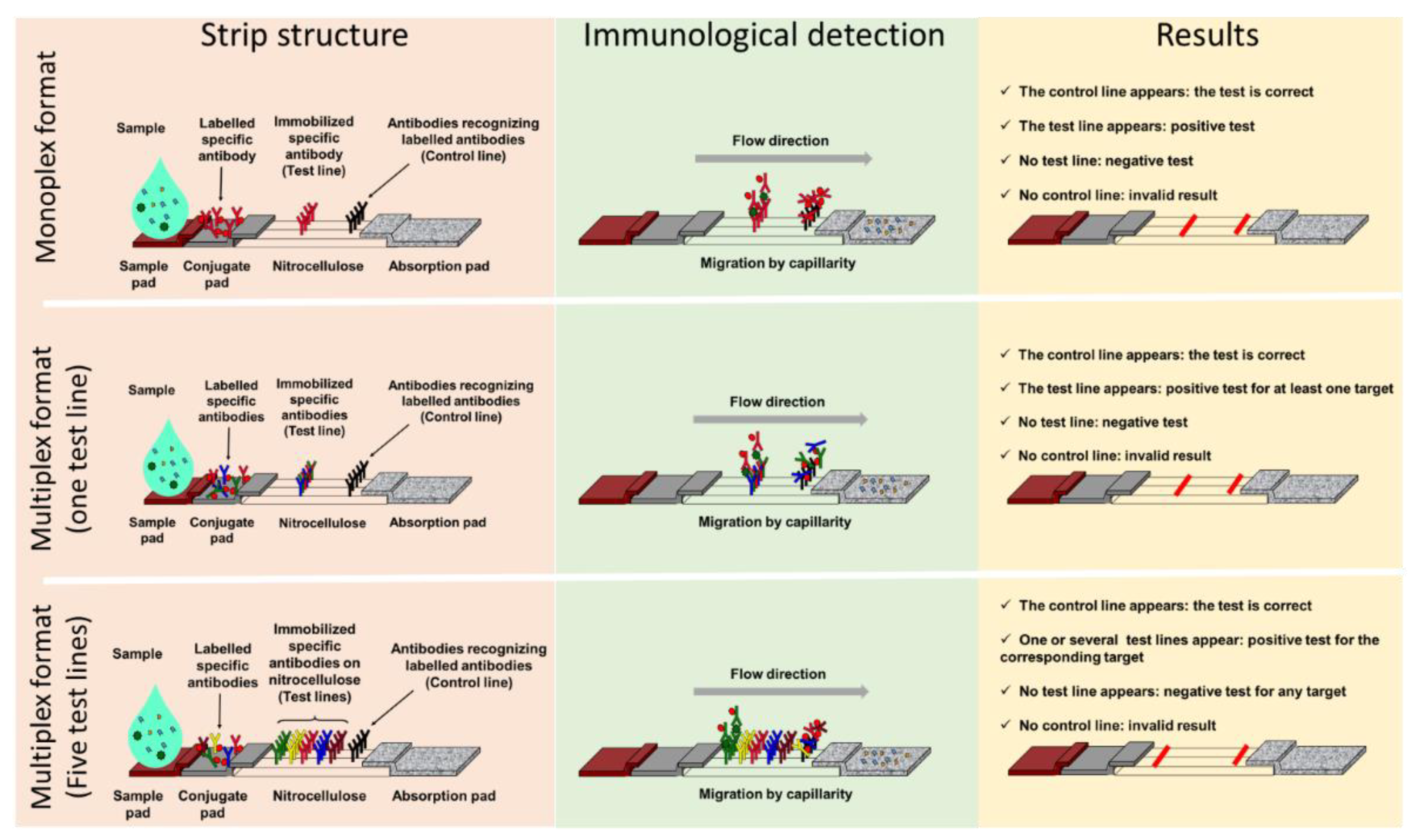
1.2. Advantages of LFIA
1.2. Advantages of Lateral Flow ImmunoAssay
2. Classical Lateral Flow Assays and AMntimicrobial Resistant
2.1. Monoplex LFIA to Address AMR Detection
2.1. Monoplex Lateral Flow ImmunoAssay to Address Antimicrobial Resistant (AMR) Detection
2.2. Multiplex LFIA in the AMR Field
2.2. Multiplex Lateral Flow ImmunoAssay in the Antimicrobial Resistant Field
2.3. Limitations of LFIA in the Context of AMR
References
- Di Nardo, F.; Chiarello, M.; Cavalera, S.; Baggiani, C.; Anfossi, L. Ten Years of Lateral Flow Immunoassay Technique Applications: Trends, Challenges and Future Perspectives. Sensors 2021, 21, 5185.
- Posthuma-Trumpie, G.A.; Korf, J.; van Amerongen, A. Lateral Flow (Immuno)assay: Its Strengths, Weaknesses, Opportunities and Threats. A Literature Survey. Anal. Bioanal. Chem. 2009, 393, 569–582.
- Park, J. An Optimized Colorimetric Readout Method for Lateral Flow Immunoassays. Sensors 2018, 18, 4084.
- Xiao, R.; Lu, L.; Rong, Z.; Wang, C.; Peng, Y.; Wang, F.; Wang, J.; Sun, M.; Dong, J.; Wang, D.; et al. Portable and Multiplexed Lateral Flow Immunoassay Reader Based on SERS for Highly Sensitive Point-of-Care Testing. Biosens. Bioelectron. 2020, 168, 112524.
- Koczula, K.M.; Gallotta, A. Lateral Flow Assays. Essays Biochem. 2016, 60, 111–120.
- Mühlberg, E.; Umstätter, F.; Kleist, C.; Domhan, C.; Mier, W.; Uhl, P. Renaissance of Vancomycin: Approaches for Breaking Antibiotic Resistance in Multidrug-Resistant Bacteria. Can. J. Microbiol. 2020, 66, 11–16.
- Kitao, T.; Miyoshi-Akiyama, T.; Shimada, K.; Tanaka, M.; Narahara, K.; Saito, N.; Kirikae, T. Development of an Immunochromatographic Assay for the Rapid Detection of AAC(6′)-Iae-Producing Multidrug-Resistant Pseudomonas Aeruginosa. J. Antimicrob. Chemother. 2010, 65, 1382–1386.
- Oshiro, S.; Tada, T.; Kameoka, Y.; Suzuki, K.; Ohmagari, N.; Miyoshi-Akiyama, T.; Kirikae, T. Development and Evaluation of Immunochromatography to Detect Gram-Negative Bacteria Producing ArmA 16S rRNA Methylase Responsible for Aminoglycoside Resistance. J. Microbiol. Methods 2015, 118, 159–163.
- Turner, N.A.; Sharma-Kuinkel, B.K.; Maskarinec, S.A.; Eichenberger, E.M.; Shah, P.P.; Carugati, M.; Holland, T.L.; Fowler, V.G. Methicillin-Resistant Staphylococcus Aureus: An Overview of Basic and Clinical Research. Nat. Rev. Microbiol. 2019, 17, 203–218.
- Yamada, K.; Wanchun, J.; Ohkura, T.; Murai, A.; Hayakawa, R.; Kinoshita, K.; Mizutani, M.; Okamoto, A.; Namikawa, T.; Ohta, M. Detection of Methicillin-Resistant Staphylococcus Aureus Using a Specific Anti-PBP2a Chicken IgY Antibody. Jpn. J. Infect. Dis. 2013, 66, 103–108.
- Amini, M.; Pourmand, M.R.; Faridi-Majidi, R.; Heiat, M.; Mohammad Nezhady, M.A.; Safari, M.; Noorbakhsh, F.; Baharifar, H. Optimising Effective Parameters to Improve Performance Quality in Lateral Flow Immunoassay for Detection of PBP2a in Methicillin-Resistant Staphylococcus Aureus (MRSA). J. Exp. Nanosci. 2020, 15, 266–279.
- Cattoir, V.; Leclercq, R. Twenty-Five Years of Shared Life with Vancomycin-Resistant Enterococci: Is It Time to Divorce? J. Antimicrob. Chemother. 2013, 68, 731–742.
- Shadel, B.N.; Puzniak, L.A.; Gillespie, K.N.; Lawrence, S.J.; Kollef, M.; Mundy, L.M. Surveillance for Vancomycin-Resistant Enterococci: Type, Rates, Costs, and Implications. Infect. Control Hosp. Epidemiol. 2006, 27, 1068–1075.
- Prematunge, C.; MacDougall, C.; Johnstone, J.; Adomako, K.; Lam, F.; Robertson, J.; Garber, G. VRE and VSE Bacteremia Outcomes in the Era of Effective VRE Therapy: A Systematic Review and Meta-Analysis. Infect. Control Hosp. Epidemiol. 2016, 37, 26–35.
- Bender, J.K.; Cattoir, V.; Hegstad, K.; Sadowy, E.; Coque, T.M.; Westh, H.; Hammerum, A.M.; Schaffer, K.; Burns, K.; Murchan, S.; et al. Update on Prevalence and Mechanisms of Resistance to Linezolid, Tigecycline and Daptomycin in Enterococci in Europe: Towards a Common Nomenclature. Drug Resist. Updates 2018, 40, 25–39.
- Oueslati, S.; Volland, H.; Cattoir, V.; Bernabeu, S.; Girlich, D.; Dulac, D.; Plaisance, M.; Laroche, M.; Dortet, L.; Simon, S.; et al. Development and Validation of a Lateral Flow Immunoassay for Rapid Detection of VanA-Producing Enterococci. J. Antimicrob. Chemother. 2021, 76, 146–151.
- Oueslati, S.; Gonzalez, C.; Volland, H.; Cattoir, V.; Bernabeu, S.; Girlich, D.; Dulac, D.; Plaisance, M.; Boutigny, L.; Dortet, L.; et al. Rapid Detection of VanA/B-Producing Vancomycin-Resistant Enterococci Using Lateral Flow Immunoassay. Diagnosis 2021, 11, 1805.
- Jacoby, G.A.; Munoz-Price, L.S. The New β-Lactamases. N. Engl. J. Med. 2005, 352, 380–391.
- Bush, K.; Jacoby, G.A. Updated Functional Classification of β-Lactamases. Antimicrob. Agents Chemother. 2010, 54, 969–976.
- Woerther, P.-L.; Burdet, C.; Chachaty, E.; Andremont, A. Trends in Human Fecal Carriage of Extended-Spectrum β-Lactamases in the Community: Toward the Globalization of CTX-M. Clin. Microbiol. Rev. 2013, 26, 744–758.
- Wareham, D.W.; Shah, R.; Betts, J.W.; Phee, L.M.; Momin, M.H.F.A. Evaluation of an Immunochromatographic Lateral Flow Assay (OXA-48 K -SeT) for Rapid Detection of OXA-48-Like Carbapenemases in Enterobacteriaceae. J. Clin. Microbiol. 2016, 54, 471–473.
- Kitao, T.; Miyoshi-Akiyama, T.; Tanaka, M.; Narahara, K.; Shimojima, M.; Kirikae, T. Development of an Immunochromatographic Assay for Diagnosing the Production of IMP-Type Metallo-β-Lactamases That Mediate Carbapenem Resistance in Pseudomonas. J. Microbiol. Methods 2011, 87, 330–337.
- Notake, S.; Matsuda, M.; Tamai, K.; Yanagisawa, H.; Hiramatsu, K.; Kikuchi, K. Detection of IMP Metallo-β-Lactamase in Carbapenem-Nonsusceptible Enterobacteriaceae and Non-Glucose-Fermenting Gram-Negative Rods by Immunochromatography Assay. J. Clin. Microbiol. 2013, 51, 1762–1768.
- Tada, T.; Sekiguchi, J.-I.; Watanabe, S.; Kuwahara-Arai, K.; Mizutani, N.; Yanagisawa, I.; Hishinuma, T.; Zan, K.N.; Mya, S.; Tin, H.H.; et al. Assessment of a Newly Developed Immunochromatographic Assay for NDM-Type Metallo-β-Lactamase Producing Gram-Negative Pathogens in Myanmar. BMC Infect. Dis. 2019, 19, 565.
- Boutal, H.; Naas, T.; Devilliers, K.; Oueslati, S.; Dortet, L.; Bernabeu, S.; Simon, S.; Volland, H. Development and Validation of a Lateral Flow Immunoassay for Rapid Detection of NDM-Producing Enterobacteriaceae. J. Clin. Microbiol. 2017, 55, 2018–2029.
- Falagas, M.E.; Kasiakou, S.K.; Saravolatz, L.D. Colistin: The Revival of Polymyxins for the Management of Multidrug-Resistant Gram-Negative Bacterial Infections. Clin. Infect. Dis. 2005, 40, 1333–1341.
- Liu, Y.-Y.; Wang, Y.; Walsh, T.R.; Yi, L.-X.; Zhang, R.; Spencer, J.; Doi, Y.; Tian, G.; Dong, B.; Huang, X.; et al. Emergence of Plasmid-Mediated Colistin Resistance Mechanism MCR-1 in Animals and Human Beings in China: A Microbiological and Molecular Biological Study. Lancet Infect. Dis. 2016, 16, 161–168.
- Ling, Z.; Yin, W.; Shen, Z.; Wang, Y.; Shen, J.; Walsh, T.R. Epidemiology of Mobile Colistin Resistance Genes Mcr-1 to Mcr-9. J. Antimicrob. Chemother. 2020, 75, 3087–3095.
- Volland, H.; Dortet, L.; Bernabeu, S.; Boutal, H.; Haenni, M.; Madec, J.-Y.; Robin, F.; Beyrouthy, R.; Naas, T.; Simon, S. Development and Multicentric Validation of a Lateral Flow Immunoassay for Rapid Detection of MCR-1-Producing Enterobacteriaceae. J. Clin. Microbiol. 2019, 57, e01454-18.
- Bernabeu, S.; Ratnam, K.C.; Boutal, H.; Gonzalez, C.; Vogel, A.; Devilliers, K.; Plaisance, M.; Oueslati, S.; Malhotra-Kumar, S.; Dortet, L.; et al. A Lateral Flow Immunoassay for the Rapid Identification of CTX-M-Producing Enterobacterales from Culture Plates and Positive Blood Cultures. Diagnostics 2020, 10, 764.
- Bianco, G.; Boattini, M.; Iannaccone, M.; Cavallo, R.; Costa, C. Evaluation of the NG-Test CTX-M MULTI Immunochromatographic Assay for the Rapid Detection of CTX-M Extended-Spectrum-β-Lactamase Producers from Positive Blood Cultures. J. Hosp. Infect. 2020, 105, 341–343.
- Nishida, S.; Nakagawa, M.; Ouchi, Y.; Sakuma, C.; Nakajima, Y.; Shimizu, H.; Shibata, T.; Kurosawa, Y.; Maruyama, T.; Okumura, C.J.; et al. A Rabbit Monoclonal Antibody-Mediated Lateral Flow Immunoassay for Rapid Detection of CTX-M Extended-Spectrum β-Lactamase-Producing Enterobacterales. Int. J. Biol. Macromol. 2021, 185, 317–323.
- Partridge, S.R.; Kwong, S.M.; Firth, N.; Jensen, S.O. Mobile Genetic Elements Associated with Antimicrobial Resistance. Clin. Microbiol. Rev. 2018, 31, e00088-17.
- Anfossi, L.; Di Nardo, F.; Cavalera, S.; Giovannoli, C.; Baggiani, C. Multiplex Lateral Flow Immunoassay: An Overview of Strategies towards High-Throughput Point-of-Need Testing. Biosensors 2018, 9, 2.
- Hamprecht, A.; Vehreschild, J.J.; Seifert, H.; Saleh, A. Rapid Detection of NDM, KPC and OXA-48 Carbapenemases Directly from Positive Blood Cultures Using a New Multiplex Immunochromatographic Assay. PLoS ONE 2018, 13, e0204157.
- Glupczynski, Y.; Evrard, S.; Huang, T.-D.; Bogaerts, P. Evaluation of the RESIST-4 K-SeT Assay, a Multiplex Immunochromatographic Assay for the Rapid Detection of OXA-48-Like, KPC, VIM and NDM Carbapenemases. J. Antimicrob. Chemother. 2019, 74, 1284–1287.
- Hong, J.; Kang, D.; Kim, D. Performance Evaluation of the Newly Developed In Vitro Rapid Diagnostic Test for Detecting OXA-48-Like, KPC-, NDM-, VIM- and IMP-Type Carbapenemases: The RESIST-5 O.K.N.V.I. Multiplex Lateral Flow Assay. Antibiotics 2021, 10, 460.
- El Kettani, A.; Maaloum, F.; Nzoyikorera, N.; Khalis, M.; Katfy, K.; Belabbes, H.; Zerouali, K. Evaluation of the Performances of the Rapid Test RESIST-5 O.O.K.N.V Used for the Detection of Carbapenemases-Producing Enterobacterales. Antibiotics 2021, 10, 953.
- Han, R.; Guo, Y.; Peng, M.; Shi, Q.; Wu, S.; Yang, Y.; Zheng, Y.; Yin, D.; Hu, F. Evaluation of the Immunochromatographic NG-Test Carba 5, RESIST-5 O.O.K.N.V., and IMP K-SeT for Rapid Detection of KPC-, NDM-, IMP-, VIM-Type, and OXA-48-like Carbapenemase Among Enterobacterales. Front. Microbiol. 2021, 11, 3296.
- Boutal, H.; Vogel, A.; Bernabeu, S.; Devilliers, K.; Creton, E.; Cotellon, G.; Plaisance, M.; Oueslati, S.; Dortet, L.; Jousset, A.; et al. A Multiplex Lateral Flow Immunoassay for the Rapid Identification of NDM-, KPC-, IMP- and VIM-Type and OXA-48-like Carbapenemase-Producing Enterobacteriaceae. J Antimicrob Chemother 2018, 73, 909–915.
- Hopkins, K.L.; Meunier, D.; Naas, T.; Volland, H.; Woodford, N. Evaluation of the NG-Test CARBA 5 Multiplex Immunochromatographic Assay for the Detection of KPC, OXA-48-Like, NDM, VIM and IMP Carbapenemases. J. Antimicrob. Chemother. 2018, 73, 3523–3526.
- Findlay, J.; Hopkins, K.L.; Meunier, D.; Woodford, N. Evaluation of Three Commercial Assays for Rapid Detection of Genes Encoding Clinically Relevant Carbapenemases in Cultured Bacteria. J. Antimicrob. Chemother. 2015, 70, 1338–1342.
- Volland, H.; Girlich, D.; Laguide, M.; Gonzalez, C.; Paris, V.; Laroche, M.; Oueslati, S.; Dortet, L.; Simon, S.; Naas, T. Improvement of the Immunochromatographic NG-Test Carba 5 Assay for the Detection of IMP Variants Previously Undetected. Antimicrob. Agents Chemother. 2019, 64, e01940-19.
- Potron, A.; Fournier, D.; Emeraud, C.; Triponney, P.; Plésiat, P.; Naas, T.; Dortet, L. Evaluation of the Immunochromatographic NG-Test Carba 5 for Rapid Identification of Carbapenemase in Nonfermenters. Antimicrob. Agents Chemother. 2019, 63, e00968-19.
- Bodendoerfer, E.; Keller, P.M.; Mancini, S. Rapid Identification of NDM-, KPC-, IMP-, VIM- and OXA-48-like Carbapenemase-Producing Enterobacteriales from Blood Cultures by a Multiplex Lateral Flow Immunoassay. J. Antimicrob. Chemother. 2019, 74, 1749–1751.
- Giordano, L.; Fiori, B.; D’Inzeo, T.; Parisi, G.; Liotti, F.M.; Menchinelli, G.; De Angelis, G.; De Maio, F.; Luzzaro, F.; Sanguinetti, M.; et al. Simplified Testing Method for Direct Detection of Carbapenemase-Producing Organisms from Positive Blood Cultures Using the NG-Test Carba 5 Assay. Antimicrob. Agents Chemother. 2019, 63, e00550-19.
- Takissian, J.; Bonnin, R.A.; Naas, T.; Dortet, L. NG-Test Carba 5 for Rapid Detection of Carbapenemase-Producing Enterobacterales from Positive Blood Cultures. Antimicrob. Agents Chemother. 2019, 63, e00011-19.
- Jenkins, S.; Ledeboer, N.A.; Westblade, L.F.; Burnham, C.-A.D.; Faron, M.L.; Bergman, Y.; Yee, R.; Mesich, B.; Gerstbrein, D.; Wallace, M.A.; et al. Evaluation of NG-Test Carba 5 for Rapid Phenotypic Detection and Differentiation of Five Common Carbapenemase Families: Results of a Multicenter Clinical Evaluation. J. Clin. Microbiol. 2020, 58, e00344-20.
- Huang, Y.-T.; Kuo, Y.-W.; Lee, N.-Y.; Tien, N.; Liao, C.-H.; Teng, L.-J.; Ko, W.-C.; Hsueh, P.-R. Evaluating NG-Test CARBA 5 Multiplex Immunochromatographic and Cepheid Xpert CARBA-R Assays among Carbapenem-Resistant Enterobacterales Isolates Associated with Bloodstream Infection. Microbiol. Spectr. 2022, 10, e01728-21.
- Liu, Z.; Bai, L.; Liu, J.; Lei, J.; Gao, X.; Tenover, F.C.; Lei, K.; Tang, Y.-W.; Geng, Y.; He, A. Parallel Validation of the NG-Test Carba 5 and the Xpert Carba-R for Detection and Characterization of Carbapenem-Resistant Enterobacterales Causing Bloodstream Infections. J. Mol. Diagn. 2021, 23, 1007–1014.
- Yoon, J.; Kim, C.H.; Yoon, S.-Y.; Lim, C.S.; Lee, C.K. Application of a Multiplex Immunochromatographic Assay for Rapid Identification of Carbapenemases in a Clinical Microbiology Laboratory: Performance and Turn-around-Time Evaluation of NG-Test Carba 5. BMC Microbiol. 2021, 21, 260.
- Zhu, Y.; Jia, P.; Li, X.; Wang, T.; Zhang, J.; Zhang, G.; Duan, S.; Kang, W.; Xu, Y.; Yang, Q. Carbapenemase Detection by NG-Test CARBA 5—a Rapid Immunochromatographic Assay in Carbapenem-Resistant Enterobacterales Diagnosis. Ann. Transl. Med. 2021, 9, 769.
- Baeza, L.L.; Pfennigwerth, N.; Greissl, C.; Göttig, S.; Saleh, A.; Stelzer, Y.; Gatermann, S.G.; Hamprecht, A. Comparison of Five Methods for Detection of Carbapenemases in Enterobacterales with Proposal of a New Algorithm. Clin. Microbiol. Infect. 2019, 25, 1286.e9–1286.e15.
- Andryukov, B.G. Six Decades of Lateral Flow Immunoassay: From Determining Metabolic Markers to Diagnosing COVID-19. AIMS Microbiol. 2020, 6, 280–304.
