2. Proline Levels Influence Pluripotency
2.1. Phenotypic Heterogeneity and Metastability of PiCs
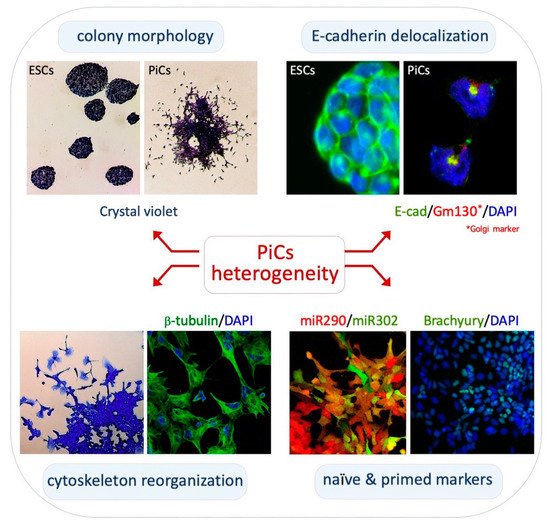
Cytoskeleton structure/networks and morphology of PiCs: PiCs are obtained upon the exogenous supplementation of L-Proline to ESCs cultured in serum/Lif (FBS/Lif) on gelatin-coated dishes
[32][33][37,38]. In these culture conditions, PiCs proliferate and give rise to highly irregular cell colonies showing three distinctive zones: a dome-shaped core of adherent naïve-like cells; a peripheral monolayer zone of polygonal/epithelial-like cells; and a crown of mesenchymal-like cells that detach from the colony (
Figure 1, top left). Naïve-like cells are round-shaped with a prominent nucleus and a reduced cytoplasm whereas detached cells display an irregular morphology with filopodia and lamellipodia protrusions
[34][39]. Staining with
β-tubulin and Vinculin antibodies and with Phalloidin shows distinctive mesenchymal features such as elongated and polarized F-actin stress fibers, frequently terminating in large and mature focal adhesion complexes (
Figure 1, bottom left)
[34][39].
Figure 1. Phenotypic heterogeneity of PiCs. Top left, crystal violet staining highlights the irregular/heterogenous structure of PiC colonies. Top right, immunofluorescence analysis showing the different localization (cell membrane vs. perinuclear structure) of E-cadherin protein (green) in naïve ESCs and motile PiCs. Bottom left, motile PiCs show a mesenchymal-like morphology with prominent filopodia and lamellipodia and a fibrillar cytoskeleton. Bottom right, expression of miR-290/mCherry and miR-302/eGFP (left) in PiCs derived from dual reporter ESCs (DRES) and immunostaining for Brachyury (right) in PiC colonies.
Furthermore, PiCs and ESCs exhibit different side scatter (SSC) and forward scatter (FSC) parameters by FACS, indicating that a high proline regimen (i.e., exogenously added proline) modifies the size and granularity of the cells
[35][40]. Of relevance, the PiCs phenotype is fully reversible; i.e., upon dissociation with accutase, PiCs fully revert to naïve ESCs when cultured in an FBS/Lif medium without proline supplementation
[34][39]. Interestingly, PiCs also revert to naïve ESCs by the addition of ascorbic acid (vitamin C; see below). This phenotypic plasticity fits with the idea that PiCs capture an intermediate state of pluripotency that can adapt to transient metabolic perturbations, in part through a reversible epigenetic mechanism (see below).
PiCs acquire a motile/invasive phenotype: Time-lapse video microscopy reveals the phenotypic ‘metastability’ of PiCs
[34][39]. Freely motile PiCs detach from the colony edges through the extension of highly dynamic and prominent protrusions and frequently divide and re-establish cell–cell adhesive contacts with neighboring motile cells or with the core of the colony
[34][39]. Thus, PiCs are prone to generate cell–cell and cell–substrate interactions in a highly dynamic and reversible manner. The ESC-to-PiC transition resembles key features of the partial epithelial-to-mesenchymal-like transition; it was thus named the embryonic stem cell-to-mesenchymal-like transition (esMT) and its reversion was named the mesenchymal-to-embryonic stem transition (MesT). Motile PiCs are able to invade matrigel in response to gradients of serum or different chemo-attractants, including Cyr61, Egf, insulin and Sdf-1, and were able to generate lung metastasis upon a tail vain injection in immunocompromised mice
[34][39]. The acquisition of mesenchymal traits and the motile/invasive features of PiCs correlate with the induction of the expression of key mesenchymal markers, including
Brachyury,
N-cadherin and
Vimentin, but not
Slug and
Snail [34][39]. Accordingly, the expression levels of the cell–cell adhesion protein E-cadherin (E-cad) are comparable between PiCs and ESCs; however, in motile PiCs, E-cad becomes delocalized from the plasma membrane and is mainly confined to intracellular vesicles; this was stained positive for trans-Golgi markers (
Figure 1, top right)
[34][39]. Accordingly, unlike that described in a canonical EMT and in the ESC-to-EpiSC transition, esMT is not associated with the transcriptional downregulation of the E-cad coding gene
Cdh1 [34][39]. In conclusion, PiCs dynamically fluctuate between two extreme phenotypes, an epithelial-like with a high tendency to preserve cell–cell interactions and a mesenchymal-like with a high cell–substrate affinity. The molecular mechanism(s) controlling the dynamic relocalization of E-cadherin during such fluctuations remains unknown and deserves further investigation.
2.2. Transcriptomic and Epigenomic Landscapes of PiCs
ESC-to-PiC transition is accompanied by extensive remodeling of the transcriptome: Proline supplementation induces the extensive remodeling of the ESC transcriptome (
Figure 2, top right), inducing the differential expression of ~1500 protein-coding genes by at least 1.5-fold
[34][39]. The expression pattern of these genes resembles in part that of FGF/Activin A (F/A)-induced EpiSCs
[34][36][39,41]. In particular, PiCs and EpiSCs share ~30% of the differentially expressed genes (DEGs). Naïve pluripotency-associated genes, including
Stella,
Pecam1,
Klf4,
Nanog,
Fbx015 and
Tbx3, are downregulated in PiCs as in EpiSCs although to a much lesser extent. Complementary to that, the priming markers
Fgf5, Brachyury, Pitx2, Otx2,
Gata6 and Foxa2 are upregulated in PiCs although less strongly than in EpiSCs. Thus, PiCs and EpiSCs share a common set of DEGs, despite their different expression levels
[34][36][39,41]. In line with the morphological and functional phenotype of PiCs, ~10% of DEGs are related to the cytoskeleton whereas ~12% correlate with cell adhesion/junction and cell motility. Moreover, ~11% of DEGs are involved in extracellular matrix (ECM) remodeling, including secreted proteases and the signaling pathways involved in cancer invasiveness such as Jak/Stat, Pi3k, MAPK and Tgfβ pathways
[34][36][39,41]. Thus, the transcriptome profile provides molecular support to the morphological and functional features of PiCs.
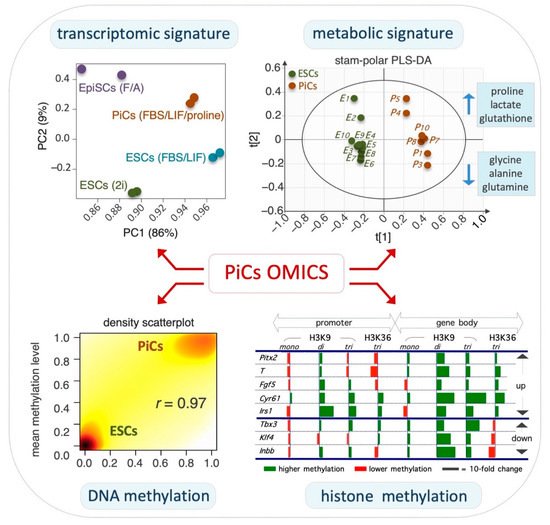
Figure 2. Global omics associated with PiC specifications.
Top left, principal component analysis of the transcriptome showing that PiCs display a gene expression profile between naïve ESCs and primed EpiSCs.
Top right, scores plot of the polar fraction of cell metabolome clearly discriminates PiCs from ESCs.
Bottom left, scatterplot showing that the global DNA methylation level is higher in PiCs compared with ESCs.
Bottom right, the relative level (PiCs vs. ESC, fold-change) of H3K9 and H3K36 histone methylation marks in selected genes. Data are modified and adapted from
[34][37][39,42].
Non-coding RNAs in the ESC-to-PiC transition: The induction of PiCs is associated with the deregulation of several classes of non-coding RNAs, including microRNAs, long non-coding RNAs (lncRNAs) and transcribed ultra-conserved elements (T-UCEs). These non-coding RNAs are emerging as key regulators of the balance between self-renewal and differentiation in pluripotent and adult stem cells
[38][39][43,44]. In this context, the ultra-conserved RNA uc.170+, also named T-UCstem1, whose expression is downregulated upon exit from naïve pluripotency
[40][41][45,46], is also deregulated in the ESC-to-PiC transition (Fico A.; personal communication). Whether uc.170+ has a functional role in the generation of PiCs remains to be further investigated.
The role of microRNAs in lineage progression has been extensively investigated. In particular, the expression profile of the microRNA clusters
miR-290/295 and
miR-302/367 is highly relevant in the transition from naïve to primed pluripotency
[42][47]. The expression of the
miR-290/295 cluster marks the naïve state and is gradually downregulated as the cells exit naïve pluripotency and progress into a primed state, which, in turn, correlates with the induction of
miR-302/367. The co-expression of
miR-290/295 and
miR-302/367 identifies an intermediate state that corresponds with an early post-implantation epiblast
[42][47]. Although naïve ESCs almost exclusively express the
miR-290/295 cluster and F/A-induced EpiSCs mainly express the
miR-302/367 cluster,
miR-290/295 and
miR-302/367 are co-expressed in PiCs (
Figure 1, bottom right), which is in line with the idea that PiCs identify an early-primed state of pluripotency
[36][41].
Epigenome remodeling in PiCs: The naïve-to-primed transition is accompanied by a global increase in DNA and histone methylation
[43][48]. Several lines of evidence indicate that this also occurs in the ESC-to-PiC transition. A time-course mass spectrometry analysis revealed that the global level of DNA 5mC progressively increases whereas that of 5hmC decreases in the ESC-to-PiC transitions (
Figure 2, bottom left)
[36][41]. Furthermore, a bisulfite-Seq (RRBSeq) analysis identified ~1.000 differentially methylated regions (DMRs) between ESCs and PiCs that are distributed throughout all chromosomes
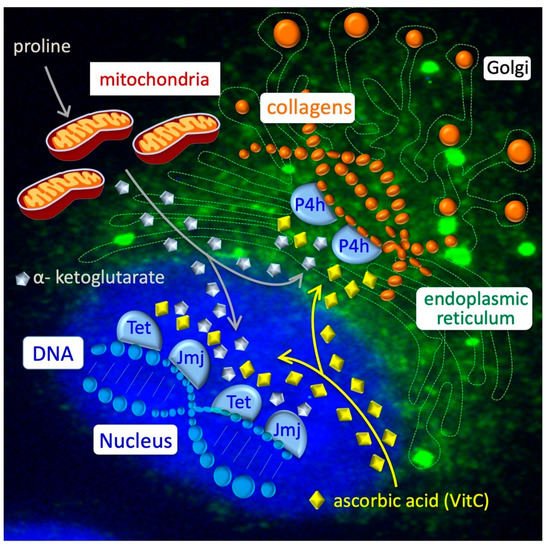
[36][41], with ~50% of them located in gene promoter regions and ~20% in gene enhancers. Of note, the majority (>90%) of hypermethylated DNA regions in PiCs were conversely hypomethylated in ESCs upon the supplementation of ascorbic acid (VitC) to the culture medium, suggesting that VitC and proline oppositely modulate methylation at the same genomic sites. It is well-known that VitC is essential for the activity of a specific class of DNA demethylases; i.e., VitC/Fe(II)/α-ketoglutarate-dependent Tet enzymes. The large majority (~90%) of hypermethylated regions in PiCs are hypermethylated as well in Tet1–3 triple knockout ESCs lacking Tet activity
[44][49], thus providing further evidence for the idea that proline and VitC oppositely regulate the activity of Tet enzymes. Another class of epigenetic enzymes that similarly require VitC for their activity is the Jmjd family of histone demethylases. The PiC specification is accompanied by a significant increase in histone H3 lysine 9 (H3K9) and histone H3 lysine 36 (H3K36) methylation levels (
Figure 2, bottom right)
[34][39]. In PiCs, H3K9me3 methylation is significantly altered at ~16.000 sites, mostly located in non-coding intergenic regions, and in ~30% of the ~1.500 DEGs
[34][39]. Conversely, H3K9me3 increases in pericentromeres and gene deserts, suggesting heterochromatin reorganization toward more dense nuclear structures
[34][39]. Of note, chromatin remodelers are released from translation inhibition by an RNA-induced silencing complex (RISC) and play a key role in the naïve-to-primed transition
[45][50]. H3K36me3 methylation is also significantly altered in ~8.000 sites in PiCs, with a large fraction (~40%) of PiC-specific genes showing significant H3K36me3 changes according to the transcriptional status
[34][39]. Together, these findings lead
reus
earchers to propose that proline supplementation interferes, either indirectly or directly, with the activity of VitC/Fe(II)/α-ketoglutarate-dependent epigenetic enzymes and this eventually results in an increase in DNA and histone methylation that accompanies the ESC-to-PiC transition (
Figure 3).
Figure 3. Collagen–epigenetic metabolic interplay in ESC-to-PiC transition. Intracellular proline is transported inside the mitochondria (grey line) and converted into α-ketoglutarate or used to load free prolyl-tRNA, which is essential for the synthesis of proline-rich proteins as collagens. Nascent collagens (orange balls) are hydroxylated in proline residues by Prolyl 4-hydroxylase (P4h) enzymes in the endoplasmic reticulum (green) and secreted through Golgi vesicles into the extracellular space. In the nucleus (blue), Tet and Jmj enzymes catalyze hydroxylation (demethylation) of DNA and histones. Reduced ascorbic acid/vitamin C (yellow diamonds) is essential for the activity of a large family of dioxygenases, including P4h, Tet and Jmj enzymes.
Several lines of evidence indicate that changes in the abundance of different metabolites could modify the epigenome in a reversible manner
[27]. Similarly, proline supplementation may reduce the availability of the metabolites/co-factors required for the activity of Tet and Jmjd enzymes, including VitC (see above;
Figure 3). It will be interesting to investigate in the future whether other epigenetic enzymes may be involved in this process, including histone acetyl transferases (HATS) and histone deacetylases (HDAC), which require acetyl-CoA
[24].