Thanks to the dawn of accessible science to map the human genome and the research that is poured into it, genetics are playing a larger role in elite sports. Genetics have a large influence over many attributes necessary for athletic excellence such as strength, muscle size, muscle fibre composition, anaerobic threshold, lung capacity, and flexibility. The aim of the study was to analyse a large database of athletes, comparing their chosen sports and the level that they play. Analysis of the individuals will include genotypes in six heavily studied genes attributed to athlete potential: ACTN3, MSTN, NOS3, ACE, AMPD1 and TRHR. A combination of both nurture and nature will always be required to bring the most out of an individual, however it would be naïve to ignore natural gifts. Genotypes may give an advantage to certain individuals but the lines between which genotype bring the most benefits is blurred, certain genotypes such as those found in MSTN are very uncommon but have very high affinity for power sports and bodybuilding. Other genes such as the once hailed “sports gene” ACTN3 have far more varied distribution and no particular athlete appeared to be hampered from any genotype, however the C allele did have affinity towards strength and power. From the six analysed genes in the study both power/strength athletes and bodybuilders had completed genotype affinity, however these genes seem to have less impact on those that compete in endurance/stamina sports. The ACTN3 C allele, MSTN G allele, NOS3 T allele, ACE II, AMPD1 C allele and TRHR C allele all show affinity towards power, strength and/or bodybuilding athletes, only the NOS3 C allele showed a true affinity towards endurance/stamina sports.
- bodybuilding
- genetics
- dna testing
- endurance
- sports
- sports performance
- athlete
- myostatin
- gene doping
1. Born Equal: Can genetics make the perfect athlete?
Introduction
Thanks to the dawn of accessible science to map the human genome and the research that is poured into it, genetics are playing a larger role in elite sports. Genetics have a large influence over many attributes necessary for athletic excellence such as strength, muscle size, muscle fibre composition, anaerobic threshold, lung capacity, and flexibility[1]. The variances (known as genotypes) in these genes affect how these genes work and therefore their physiological input. Whilst nurturing talent is of the utmost importance, we considered how much importance genotypes play in an athlete being attracted to a certain sport and how much influence they can bring to performance. This is a preliminary study analysing specific genotypes known to have influence over athletic traits on a large database of 4477.
Aim
The aim of the study was to analyse a large database of athletes (N= 4477), comparing their chosen sports and the level that they play. Analysis of the individuals will include genotypes in six heavily studied genes[2] attributed to athlete potential: ACTN3, MSTN, NOS3, ACE, AMPD1 and TRHR. These results will be collated to see if there are potential correlations between certain genotypes, the sport played, and the levels played.
Methods
The subject’s data is anonymously pulled from the Muhdo health ltd. database and includes individuals who have chosen that they are athletes through an in-app questionnaire, the term "athlete" also includes those that use the gym on a regular basis. There are 4447 individuals in the database currently under the status of being an athlete. 1876 female stated as female and 2601 stated as male, they are of varied heritage and have an age range of 23 - 52 years.
Muhdo Health utilise array technology, which is a custom created array from Illumina, this array analyses 1000 genotypes associated with health, performance, and diet. Phosphate buffered saline was used to prevent any premature cell rupture and cell recovery is accomplished using standard extraction procedures. Fluorescence information is analysed and standardised into Illumina’s GenomeStudio®. The genotypes chosen for this analysis iScan Control Software automatically normalized the intensity of the fluorescence data to remove technical variation and generated genotype calls. Three probes were used in different orientations and repeated three times to ensure accurate readings and calls of the alleles. Nucleotides (Adenine, Thymine, Guanine and Cytosine) were listed in the forward-forward orientation, with a reported 99.9% accuracy. If this information was not consistent in all three calls the allele set was not included for the subsequent statistical analysis.
The genotypes chosen for this analysis are as follows:
Table 1. Genotypes analysed.
|
rs1815739 (ACTN3) |
|
CC |
|
CT |
|
TT |
|
rs1805086 (MSTN) |
|
AA |
|
AG |
|
GG |
|
rs2070744 (NOS3) |
|
CC |
|
CT |
|
TT |
|
rs4343 (ACE) |
|
II |
|
ID |
|
DD |
|
rs17602729 (AMPD1) |
|
CC |
|
CT |
|
TT |
|
rs16892496 (TRHR) |
|
CC |
|
AC |
|
AA |
Athletes are put into certain groups based on the sports they state they play:
Table 2. Sports associated with individuals.
|
Sports association in group (STRENGTH/POWER) |
Sports association in group (ENDURANCE/STAMINA) |
Sports association in group (BODYBUILDING) |
|
Sprinting - 100 - 400m |
Long distance running |
Resistance training for aesthetic events/look (bodybuilding, physique) |
|
Powerlifting |
Cycling |
|
|
Olympic lifting |
Tennis |
|
|
Long Jump |
Rowing |
|
|
Triple Jump |
Football/Soccer |
|
|
Olympic throw events |
||
|
Sprint cycling |
||
|
Resistance training for primary strength increases |
||
|
Sports association in group (STRENGTH/POWER) N = 1981 |
Sports association in group (ENDURANCE/STAMINA) N = 1381 |
Sports association in group (BODYBUILDING) N = 1115 |
|
Sprinting - 100 - 400m |
Long distance running |
Resistance training for aesthetic events/look (bodybuilding, physique) |
|
Powerlifting |
Cycling |
|
|
Olympic lifting |
Tennis |
|
|
Long Jump |
Rowing |
|
|
Triple Jump |
Football/Soccer |
|
|
Olympic throw events |
||
|
Sprint cycling |
||
|
Resistance training for primary strength increases |
2. Literature review
ACTN3
The ACTN3 gene encodes the alpha-actinin-3 protein, a structural component of the Z-disc that anchors actin-containing thin filaments and contributes to regulating muscle length and tension during contraction. Alpha-actinin-3 is only expressed in type 2 muscle fibres [3] . A common polymorphism, R577X (rs1815739), of the “speed gene” involves a C-to-T base substitution resulting in an arginine residue being replaced with a premature stop codon. TT homozygotes have been shown to have reduced alpha-actinin-3 protein expression and a reduced proportion of fast twitch muscle fibres [3]. R577X is associated with strength and power, and it has been widely demonstrated that TT homozygotes have impaired ability to produce powerful contractions and reduced sprint and power performance and that there is a lower prevalence of TT homozygotes amongst elite athletes [4]. Furthermore, a meta-analysis of published studies corroborated the association between the C allele and elite power athletes [5]. There is also emerging evidence that in addition to increased strength and power potential, the C allele has a positive impact on training response, exercise recovery and injury risk [6]. There have also been studies suggesting the T allele is slightly advantageous for endurance sports, but the results are not unequivocal and there is a low prevalence in highly successful Kenyan and Ethiopian endurance runners [7].
MSTN
The MSTN gene encodes the myostatin protein of the TGF-β protein superfamily. Myostatin is a negative regulator of muscle cell growth and differentiation ensuring the muscles don’t grow too large and is also involved in regulation of adipose tissue [8]. Loss of function MSTN mutations in animals with very high skeletal muscle mass, a phenomenon known as double muscling, have been observed [9] The evidence indicating the role myostatin plays in skeletal muscle regulation has led to WADA prohibiting the use of myostatin inhibitors and labelled research into their use an area of concern [10]. Additionally, MSTN -/- mice were shown to have increased number and size of skeletal muscle fibres [9]. The K153R A-to-G base substitution (rs1805086) has previously been associated with an increased hypertrophic response to strength training [11]. The G allele frequency has been estimated to be ~7% (3% in Caucasians, 22% in Africans) and some studies conducted so far have failed to find a significant association between the variant and strength and power due to this low frequency and limited sample size [12]. Furthermore, myostatin levels have been shown to be proportional to age and sex leading to some contradictory findings [13]. Despite this, a meta-analysis by Olegovich et al of the K153R polymorphism data combined from 4 studies found a statistically significant difference in the prevalence of the G allele in athletes in weightlifting sports vs control groups. The findings of 3 of the 4 studies was not significant due to the low frequency of the allele and limited sample number [12].
NOS3
The NOS3 gene encodes the Nitric Oxide Synthase 3 protein that catalyses the synthesis of the potent endothelium derived relaxation factor Nitric Oxide (NO). NO regulates skeletal muscle Oxygen uptake. The 786 T/C polymorphism (rs2070744) has been associated with several physical performance traits including exercise-induced response to hypoxia, aerobic capacity, and heart and vascular responses [14]. Furthermore, the SNP has been associated with both power and endurance athletes with the T allele being significantly more prevalent in elite power athletes compared to controls [15].
ACE
ACE is a gene that encodes the angiotensin converting enzyme, a protein that is part of the renin-angiotensin system that regulates blood pressure and the balance of salt and fluids in the body. ACE raises blood pressure via several pathways: conversion of angiotensin I into angiotensin II, a vasoconstrictor; promotion of aldosterone that induces reabsorption of salt and fluids in the kidneys; and inactivation of bradykinin, a vasodilator. ACE has 2 main alleles: the I allele with a 287bp insertion; and the D allele without the 287bp. The I allele has reduced enzymatic activity [16]. The I allele has been widely demonstrated to be more prevalent amongst endurance athletes versus controls [16]. A meta-analysis of previous published studies corroborated these findings by identifying a significant association between II homozygosity and endurance athletes. However, the I allele was just outside the threshold for significance [5]. Additionally, the D allele has been associated with strength and power athletes [17].
AMPD1
The adenosine monophosphate deaminase enzyme encoded by AMPD1 is an important regulator of skeletal muscle energy metabolism. During physical activity the enzyme converts AMP into inosine monophosphate (IMP) as part of the purine nucleotide cycle, releasing an ammonia molecule and providing energy for the skeletal muscle. The rs17602729 C-to-T Gln12X variant is associated with reduced AMPD activity and is less prevalent in power athletes than controls [18] .
TRHR
TRHR encodes the thyrotropin releasing hormone receptor. Thyrotropin releasing hormone binding induces the release of thyrotropin and prolactin from the anterior pituitary gland and thereby affecting the body’s metabolism and various hormonal activities including milk production. The rs16892496 A allele is associated with TRH resistance and increased lean body mass [19] . Additionally, the T allele in rs7832552 of TRHR has been associated with elite sprinters [20].
23. Results
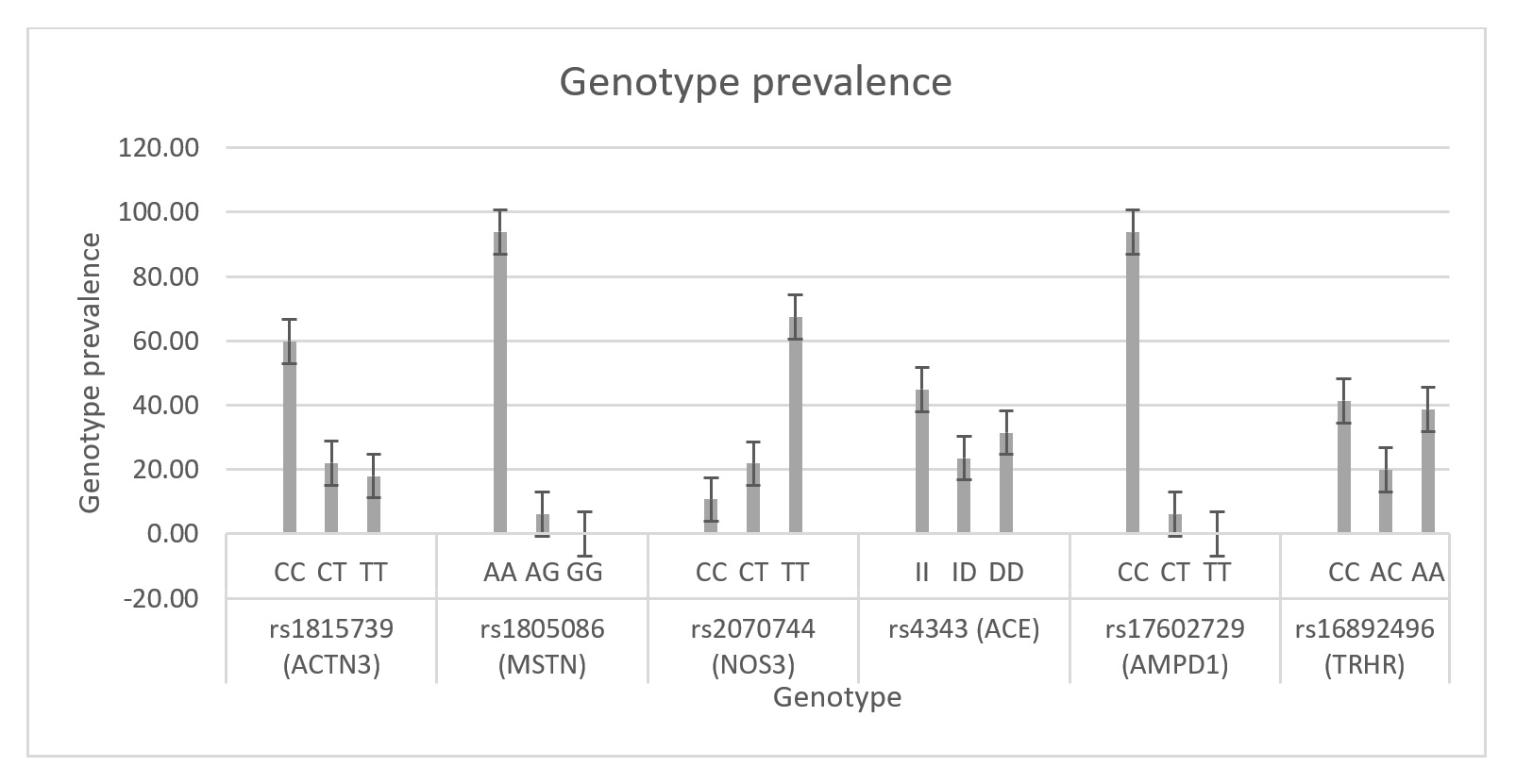
After analysis of the results the following was found: (if 75% or above of individuals were attributed to specific sports this would be stated as “predominate” if there was over 90% this would be stated outright)
Table 3. Results.
|
Power/Strength Talent Bias |
Endurance/Stamina Bias |
Bodybuilder Bias |
|
|
ACTN3 - CC |
ACTN3 - ANY |
ACTN3 - CC |
|
|
MSTN - AG/GG |
MSTN - ANY, (maybe) AA |
MSTN - AG/GG |
|
|
NOS3 - TT |
NOS3 - CC |
NOS3 - (maybe) TT |
|
|
ACE - II |
ACE - ANY |
ACE - (maybe) II |
|
|
AMPD1 - CC/CT |
AMPD1 - (maybe) TT. CC/CT |
AMPD1 - CC/CT |
|
|
TRHR - CC |
TRHR - ANY |
TRHR - CC | |
|
rs1815739 (ACTN3) |
Subjects |
Sports |
Notes |
|
CC |
2683 |
All/Predominate power/Bodybuilding (2087 - 77.79%) |
|
|
CT |
988 |
All |
|
|
TT |
806 |
All |
|
|
rs1805086 (MSTN) |
|||
|
AA |
4205 |
All |
|
|
AG |
271 |
Power/Strength/Bodybuilding (254 - 93.7%) |
32/254 compete at national/international level in strength/bodybuilding/power |
|
GG |
1 |
Power/Strength (100%) |
2x WSM |
|
rs2070744 (NOS3) |
|||
|
CC |
480 |
Endurance/Stamina (433 - 90.21%) |
|
|
CT |
979 |
All |
|
|
TT |
3018 |
All/Predominate power (2380 - 78.8%) |
123/2380 compete at national level or higher in power sports |
|
rs4343 (ACE) |
|||
|
II |
2011 |
Power (1890 - 93.98%) |
|
|
ID |
1056 |
All |
|
|
DD |
1410 |
All |
|
|
rs17602729 (AMPD1) |
|||
|
CC |
4200 |
All |
2700 compete at a national level or international level |
|
CT |
275 |
All |
|
|
TT |
2 |
Endurance/Stamina (100%) |
2/2 compete at national level |
|
rs16892496 (TRHR) |
|||
|
CC |
1856 |
Power/strength/bodybuilding (1760 - 94.83%) |
|
|
AC |
893 |
All |
|
|
AA |
1728 |
All |
FResults show that the prevalence of ceromtain genotypes in the analysed data there are some clear thlete population is varied. The “C” allele in ACTN3 (rs1815739) is overrepresented in the whole athlete population making up 60% of this population, however this steams primarily from strength/bodybuilder athletes. The “G” allele in MSTN (rs1805086) is uncommon in any population and this confirms the rarity, the common genotypes may not play a role in achieving elite success however the “G” allele is highly associated with certain strength and bodybuilding. The “T” allele in NOS3 (rs2070744) is overrepresented in power sports whilst others are far vaguer. Fromand is also overrepresented in the whole athlete population, the “C” allele may be associated with endurance and stamina sports. The II variance in the ACE (rs4343) gene is overrepresented across the above data alone, the following genotypes maybe associated with specific sports:thlete population and may show a higher affinity towards power sports. The “C” allele in AMPD1 (rs17602729) is highly represented in the athlete population which matches population studies. The “C” allele in TRHR (rs16892496) shows higher affinity for strength/bodybuilding sports but the “A” allele does not appear to cause any deficit for performance.
Table 4. Sport specific genotypes.
101/1760 compete at national level in bodybuilding | ||
|
Power/Strength Talent Bias |
Endurance/Stamina Bias |
Bodybuilder Bias |
|
ACTN3 - CC |
ACTN3 - ANY |
ACTN3 - CC |
|
MSTN - AG/GG |
MSTN - ANY, (maybe) AA |
MSTN - AG/GG |
|
NOS3 - TT |
NOS3 - CC |
NOS3 - (maybe) TT |
|
ACE - II |
ACE - ANY |
ACE - (maybe) II |
|
AMPD1 - CC/CT |
AMPD1 - (maybe) TT. CC/CT |
AMPD1 - CC/CT |
|
TRHR - CC |
TRHR - ANY |
TRHR - CC |
TGraph 1. Genotype prevable 4ence.
3. Discussion
ACTN3
ACTN3 and its single nucleotide polymorphism R577X (rs1815739). R577X directly determines the expression of the α-actinin-3 protein that contributes to the construction of the contractile component in power-generating fast twitch fibres of skeletal muscle. A number of association studies have demonstrated that ACTN3 is a strong candidate in the influence of elite athletic performance[321]. Most studies attributed the C allele to power sports and the T allele to endurance sports[321]. In this study the C allele was associated with strength and power sports and was also represented higher in the bodybuilder category, however the T allele was not associated directly with endurance or stamina sports and therefore there is no candidate variance of ACTN3 associated with endurance and stamina sports.
MSTN
MSTN is a gene that makes instructions for producing the protein myostatin, a protein that is part of the transforming growth factor beta family (TGFβ). The TGFβ family of proteins control the growth of tissues in the body, myostatin is found nearly exclusively in the skeletal muscles where it is active before and after birth. The protein actually controls skeletal growth by restraining it, preventing muscles becoming excessively large[422]. Current research that surrounds myostatin is based around its potential treatment in muscle wasting disorders, animals that have mutations in the encoding gene MSTN show greater muscle mass, strength and in some circumstances reduced bodyfat, which can be known as myostatin-related muscle hypertrophy[523]. The current study shows that the majority of athletes have the common AA variance in rs1805086 and therefore it is unlikely to affect athletic potential for the majority. However those who are heterozygotes are nearly completely associated with strength, power or bodybuilding with a high level of these competing at a high level in their given sports. There was only one individual with the complete mutation within the gene and this individual competes at the highest level in strength-based sports. It can be stated the G allele is associated with strength, power and bodybuilding which would match the current research on myostatin.
NOS3
The NOS3 gene encodes an enzyme which is responsible for the production of the small molecule nitric oxide (NO). NOS3 is predominantly expressed in the endothelial tissue which lines the circulatory system and heart, where it plays a key role in regulation of NO. NO is a vasodilator and aids the cardiovascular system[624] when produced correctly, and this vasodilation effect is attributed to an increase in sporting performance through increasing oxygen and other nutrients to muscles. This study shows that the C allele is associated with endurance and stamina and the T allele is associated with strength and power.
ACE
ACE encodes the enzyme angiotensin-converting enzyme, this enzyme is able to cleave proteins. It helps regulate blood pressure and balance fluids and salts in the body. The ACE II genotype has been associated with muscle damage and the DD genotype with protective properties[725], it is hypothesised that muscle damage can lead to greater adaptation. The II genotype is often attributed to athlete status, however in this study the II genotype was primarily associated with strength and power athletes only.
AMPD1
Adenosine monophosphate deaminase 1 (AMPD1) plays a vital role in the purine nucleotide cycle, the gene encodes an enzyme of the same name. The enzyme coverts adenosine monophosphate to inosine monophosphate which frees an ammonia molecule during the process. Mutations that occur within the AMPD1 gene are one of the most common defects detected in the Caucasian population with a likelihood of having the mutations as 1-2%. Several studies indicate that certain variants can cause fatigue, muscle weakness and muscular cramps, however some even with these variants remain asymptomatic[826].
The disorder caused by mutations is known as adenosine monophosphate deaminase deficiency type 1 (AMPD1 deficiency) or myoadenylate deaminase deficiency (MADD). The most common symptoms of AMPD1 deficiency are:
- Exercise intolerance – symptoms of fatigue and fast onset weakness on the commencement of exertion or prolonged exertion.
- Fatigue – general fatigue is poorly understood and may have multiple pathways; however a surplus of adenosine reduces alertness.
- Muscle cramping – this may be due to an increased lactate.
Based on the genotypes effect on the body we would expect that those with the T allele would suffer from poor exercise performance. However, this study shows no affect on athlete profile and the genotypes in AMPD1. It should be stated that most athletes had the non-mutated genotypes and for power and strength sports especially it could be stated that at least one C allele is beneficial. Only two individuals had TT and both were endurance/stamina athletes. It is probable that the C allele is beneficial for all sports, especially for power and strength sports and all variances are applicable for endurance and stamina sports.
TRHR
Thyrotropin releasing hormone receptor is a gene that is not as well analysed as the other five in the study. Through a chemical cycle within the body the gene may alter the levels of thyroid-stimulating (TSH) in the blood[927]. The genotype analysed in the study rs16892496 appears to affect lean body mass[1019], with the C allele causing an increase in lean body mass. In the study the C allele is heavily associated with power and strength sports whereas it is not associated with endurance and stamina sports.
4. Conclusion
TMany of the studies conducted previously concur by identifying significant allele-sport type associations. However some published papers appear to contradis is a preliminary stuct these studies by not finding any significant differences in allele prevalence between elite athletes and control groups. However, it is important to note that many previous studies have limited sample size and therefore lack statistical power. Furthermore, given the rarity of many of the minor alleles there has not been sufficient allele representation to achieve statistically significant results. This study aiming to ins to help rectify the paucity of studies with large study groups and to the author’s knowledge this is the highest sample size to date in a study looking at associations between polymorphisms and athlete status and physical performance.
4. Conclusion
The study aimed to investigate the database of Muhdo Health ltd. The primary reasoning is the lack of large genetic database studies being conducted largely on the athlete population. The findings in this study show that there may be a genetic blueprint or profile for certain athletes. Many more genotypes need to be analysed in further study to create a larger picture of how they work together to cause advantages for certain sports, one limitation of the study may be the inclusion of athletes who regularly frequent the gym but who would not otherwise be classified as elite athletes.
A combination of both nurture and nature will always be required to bring the most out of an individual, however it would be naïve to ignore natural gifts. Genotypes may give an advantage to certain individuals but the lines between which genotype bring the most benefits is blurred, certain genotypes such as those found in MSTN are very uncommon but have very high affinity for power sports and bodybuilding. Other genes such as the once hailed “sports gene” ACTN3[1] have far more varied distribution and no particular athlete appeared to be hampered from any genotype, however the C allele did have affinity towards strength and power. From the six analysed genes in the study both power/strength athletes and bodybuilders had completed genotype affinity, however these genes seem to have less impact on those that compete in endurance/stamina sports. The ACTN3 C allele, MSTN G allele, NOS3 T allele, ACE II, AMPD1 C allele and TRHR C allele all show affinity towards power, strength and/or bodybuilding athletes, only the NOS3 C allele showed a true affinity towards endurance/stamina sports.
References
- Lee Billings; The Sports Gene. Scientific American 2013, 309, 96-96, 10.1038/scientificamerican0813-96d.
- Dov Greenbaum; Jieming Chen; Mark Gerstein; Deep Inside Champions, Just Genes? The Sports Gene Inside the Science of Extraordinary Athletic Performance/What Makes the Perfect Athlete by David Epstein Current, New York, 2013. 352 pp. $26.95. ISBN 9781591845119. Yellow Jersey, London. £16.99. ISBN 978-0224091619.. Science 2013, 342, 560-561, 10.1126/science.1245795.
- Stephen M Roth; Sean Walsh; Dongmei Liu; E Jeffrey Metter; Luigi Ferrucci; Ben F Hurley; The ACTN3 R577X nonsense allele is under-represented in elite-level strength athletes. Daniel G. MacArthur; Kathryn N. North; ACTN3. Euxeropean Journal of Human Geneticcise and Sport Sciences Reviews 2007, 16, 391-394, 10.1038/sj.ejhg.5201964., 35, 30-34, 10.1097/jes.0b013e31802d8874.
- Gilles Carnac; Stéphanie Ricaud; Barbara Vernus; Anne Bonnieu; Myostatin: biology and clinical relevance.. Mini-RNan Yang; Daniel G. MacArthur; Jason P. Gulbin; Allan G. Hahn; Alan H. Beggs; Simon Easteal; Kathryn North; ACTN3 Genotype Is Associated with Human Elite Athletic Performance. Thevi Amews in Medicinal Chemirican Journal of Human Geneticstry 2006, 6, 765-770, 10.2174/138955706777698642.3, 73, 627-631, 10.1086/377590.
- J.A. Stockman; Myostatin Mutation Associated With Gross Muscle Hypertrophy in a Child. YearbookFang Ma; Yu Yang; Xiangwei Li; Feng Zhou; Cong Gao; Mufei Li; Lei Gao; The Association of Sport Performance with ACE and ACTN3 Genetic Polymorphisms: A Systematic Review and Meta-Analysis. PLOS of Pediatrics ONE 2006, 2006, 319-321, 10.1016/s0084-3954(07)70193-8.13, 8, e54685, 10.1371/journal.pone.0054685.
- Khalid M Naseem; The role of nitric oxide in cardiovascular diseases. MCraig Pickering; John Kiely; ACTN3: More than Just a Gene for Speed. Frolntiecular Aspects of Medicine rs in Physiology 2005, 26, 33-65, 10.1016/j.mam.2004.09.003.17, 8, 1080, 10.3389/fphys.2017.01080.
- Chen Yamin; Offer Amir; Moran Sagiv; Eric Attias; Yoav Meckel; Nir Eynon; Michael Sagiv; Ruthie E. Amir; ACE ID genotype affects blood creatine kinase response to eccentric exercise. JourNan Yang; Daniel G. Macarthur; Bezabhe Wolde; Vincent O. Onywera; Michael K. Boit; Sau Yin Mary-Ann Lau; Richard H. Wilson; Robert A. Scott; Yannis P. Pitsiladis; Kathryn North; et al. The ACTN3 R577X Polymorphism in East and West African Athletes. Medicinale of Applied Physiology & Science in Sports & Exercise 2007, 10, 3, 2057-2061, 10.1152/japplphysiol.00867.2007.9, 1985-1988, 10.1249/mss.0b013e31814844c9.
- Rolf Kreutz; Rayan Saab; Ronald Mastouri; ADENOSINE MONOPHOPHATE DEAMINASE-1 (AMPD1) DEFICIENCY AND RESPONSE TO REGADENOSON. JoBing Deng; Feng Zhang; Jianghui Wen; Shengqiang Ye; Lixia Wang; Yu Yang; Ping Gong; Siwen Jiang; The function of myostatin in the regulation of fat mass in mammals. Nutrnal of the American College of Cardiology ition & Metabolism 2014, 63, A1113, 10.1016/s0735-1097(14)61113-x.7, 14, 1-6, 10.1186/s12986-017-0179-1.
- Mario Mellado; Teresa Fernández-Agulló; Jose Miguel Rodriguez Frade; Miriam García San Frutos; Pilar De-La-Pena-Cortines; Carlos Martı́nez-A; Eladio Montoya; Expression analysis of the thyrotropin-releasing hormone receptor (TRHR) in the immune system using agonist anti-TRHR monoclonal antibodies. FEBSAlexandra C. McPherron; Se-Jin Lee; Double muscling in cattle due to mutations in the myostatin gene. Proceedings of Lthetter National Academy of Sciences 1999, 7, 9451, 308-314, 10.1016/s0014-5793(99)00607-9., 12457-12461, 10.1073/pnas.94.23.12457.
- Xiao-Gang Liu; Li-Jun Tan; Shu-Feng Lei; Yong-Jun Liu; Hui Shen; Liang Wang; Han Yan; Yan-Fang Guo; Dong-Hai Xiong; Xiang-Ding Chen; et al.Feng PanTie-Lin YangYin-Ping ZhangNelson L. TangXue-Zhen ZhuHong-Yi DengShawn LevyRobert R. ReckerChristopher J. Papasian Genome-wide Association and Replication Studies Identified TRHR as an Important Gene for Lean Body Mass. The American Journal of Human Genetics 2009, 84, 418-423, 10.1016/j.ajhg.2009.02.004.Prohibited Substances . Wadaama. Retrieved 2022-8-13
- Matthew A. Kostek; Theodore J. Angelopoulos; Priscilla M. Clarkson; Paul M. Gordon; Niall M. Moyna; Paul S. Visich; Robert F. Zoeller; Thomas B. Price; Richard L. Seip; Paul D. Thompson; et al.Joseph M. DevaneyHeather Gordish-DressmanEric P. HoffmanLinda S. Pescatello Myostatin and Follistatin Polymorphisms Interact with Muscle Phenotypes and Ethnicity. Medicine & Science in Sports & Exercise 2009, 41, 1063-1071, 10.1249/mss.0b013e3181930337.
- Aksenov Maxim Olegovich; Marek Kruszewski; Association of Myostatin Gene Polymorphisms with Strength and Muscle Mass in Athletes: a Systematic Review and Meta-Analysis of the MSTN rs1805086 Mutation. null 2022, 1, 1, 10.21203/rs.3.rs-1342135/v1.
- Michael J. Seibert Ms; Qian-Li Xue; Linda P. Fried; Jeremy D. Walston; Polymorphic Variation in the Human Myostatin (GDF-8) Gene and Association with Strength Measures in the Women's Health and Aging Study II Cohort. Journal of the American Geriatrics Society 2001, 49, 1093-1096, 10.1046/j.1532-5415.2001.49214.x.
- Piotr Zmijewski; Paweł Cięszczyk; Ildus I. Ahmetov; Piotr Gronek; Ewelina Lulinska; Marcin Dornowski; Agata Rzeszutko; Jakub Chycki; Waldemar Moska; Marek Sawczuk; et al. The NOS3 G894T (rs1799983) and -786T/C (rs2070744) polymorphisms are associated with elite swimmer status. Biology of Sport 2018, 35, 313-319, 10.5114/biolsport.2018.76528.
- Félix Gómez-Gallego; Jonatan R. Ruiz; Amaya Buxens; Marta Artieda; David Arteta; Catalina Santiago; Gabriel Rodríguez-Romo; José I. Lao; Alejandro Lucia; The −786 T/C polymorphism of the NOS3 gene is associated with elite performance in power sports. Graefe's Archive for Clinical and Experimental Ophthalmology 2009, 107, 565-569, 10.1007/s00421-009-1166-7.
- Zudin Puthucheary; James R. A. Skipworth; Jai Rawal; Mike Loosemore; Ken Van Someren; Hugh E. Montgomery; The ACE Gene and Human Performance. Sports Medicine 2011, 41, 433-448, 10.2165/11588720-000000000-00000.
- J. Eider; P. Cieszczyk; K. Ficek; A. Leonska-Duniec; M. Sawczuk; A. Maciejewska-Karlowska; A. Zarebska; The association between D allele of the ACE gene and power performance in Polish elite athletes. Science & Sports 2013, 28, 325-330, 10.1016/j.scispo.2012.11.005.
- O. N. Fedotovskaya; A. A. Danilova; I. I. Akhmetov; Effect of AMPD1 Gene Polymorphism on Muscle Activity in Humans. Bulletin of Experimental Biology and Medicine 2013, 154, 489-491, 10.1007/s10517-013-1984-9.
- Xiao-Gang Liu; Li-Jun Tan; Shu-Feng Lei; Yong-Jun Liu; Hui Shen; Liang Wang; Han Yan; Yan-Fang Guo; Dong-Hai Xiong; Xiang-Ding Chen; et al.Feng PanTie-Lin YangYin-Ping ZhangNelson L. TangXue-Zhen ZhuHong-Yi DengShawn LevyRobert R. ReckerChristopher J. Papasian Genome-wide Association and Replication Studies Identified TRHR as an Important Gene for Lean Body Mass. The American Journal of Human Genetics 2009, 84, 418-423, 10.1016/j.ajhg.2009.02.004.
- Eri Miyamoto-Mikami; Haruka Murakami; Hiroyasu Tsuchie; Hideyuki Takahashi; Nao Ohiwa; Motohiko Miyachi; Takashi Kawahara; Noriyuki Fuku; Lack of association between genotype score and sprint/power performance in the Japanese population. Journal of Science and Medicine in Sport 2016, 20, 98-103, 10.1016/j.jsams.2016.06.005.
- Stephen M Roth; Sean Walsh; Dongmei Liu; E Jeffrey Metter; Luigi Ferrucci; Ben F Hurley; The ACTN3 R577X nonsense allele is under-represented in elite-level strength athletes. European Journal of Human Genetics 2007, 16, 391-394, 10.1038/sj.ejhg.5201964.
- Gilles Carnac; Stéphanie Ricaud; Barbara Vernus; Anne Bonnieu; Myostatin: biology and clinical relevance.. Mini-Reviews in Medicinal Chemistry 2006, 6, 765-770, 10.2174/138955706777698642.
- J.A. Stockman; Myostatin Mutation Associated With Gross Muscle Hypertrophy in a Child. Yearbook of Pediatrics 2006, 2006, 319-321, 10.1016/s0084-3954(07)70193-8.
- Khalid M Naseem; The role of nitric oxide in cardiovascular diseases. Molecular Aspects of Medicine 2005, 26, 33-65, 10.1016/j.mam.2004.09.003.
- Chen Yamin; Offer Amir; Moran Sagiv; Eric Attias; Yoav Meckel; Nir Eynon; Michael Sagiv; Ruthie E. Amir; ACE ID genotype affects blood creatine kinase response to eccentric exercise. Journal of Applied Physiology 2007, 103, 2057-2061, 10.1152/japplphysiol.00867.2007.
- Rolf Kreutz; Rayan Saab; Ronald Mastouri; ADENOSINE MONOPHOPHATE DEAMINASE-1 (AMPD1) DEFICIENCY AND RESPONSE TO REGADENOSON. Journal of the American College of Cardiology 2014, 63, A1113, 10.1016/s0735-1097(14)61113-x.
- Mario Mellado; Teresa Fernández-Agulló; Jose Miguel Rodriguez Frade; Miriam García San Frutos; Pilar De-La-Pena-Cortines; Carlos Martı́nez-A; Eladio Montoya; Expression analysis of the thyrotropin-releasing hormone receptor (TRHR) in the immune system using agonist anti-TRHR monoclonal antibodies. FEBS Letters 1999, 451, 308-314, 10.1016/s0014-5793(99)00607-9.
