Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Jianping Wu and Version 2 by Conner Chen.
The Na+/H+ exchangers (NHEs) are membrane transporters that exchange one intracellular pro-ton for one extracellular Na+. The first discovered NHE isoform, NHE1, is expressed almost ubiquitously in all tissues, especially in the myocardium. During myocardial ische-mia-reperfusion, NHE1 catalyzes increased uptake of intracellular Na+, which in turn leads to Ca2+ overload and subsequently myocardial injury. Numerous preclinical research has shown that NHE1 is involved in cardiac hypertrophy and heart failure, but the exact molecular mecha-nisms remain elusive. The objective of this review is to demonstrate the potential role of NHE1 in cardiac hypertrophy and heart failure and investigate the underlying mechanisms.
- Na+/H+ exchanger 1
- cardiac hypertrophy
- heart failure
1. Distribution and Structure of NHE1
1.1. NHE1 Distribution
Mitchell et al. [1] were the first to hypothesize the presence of an electroneutral transport mechanism for cation and H+ to exchange across the mitochondrial inner membrane. Subsequently, several studies verified the activity of NHEs in the bacterium, which was later described in mammals [2].
NHEs are encoded by the SLC9 gene family of the solute carrier (SLC) transporters [3]. Three subfamilies of the SLC9 gene family have been identified based on phylogenetic analysis. The SLC9A subgroup comprises nine mammalian NHE paralogs, NHE1-9. The SLC9B subgroup includes two Na+ or Li+/H+ antiporters, NHA1 (SLC9B1) and NHA2 (SLC9B2). The SLC9C subgroup consists of NHE10 (SLC9C1), expressed primarily in sperm tissue and osteoblast, and an orphan-related protein NHE11 (SLC9C2) [2][3][4][5].
The members of the mammalian SLC9A gene family are divided into plasma membrane clusters and intracellular organelle clusters. The NHE family members NHE1-5 are primarily found in the plasma membranes, while NHE6-9 localize to the organelle chambers [6]. Among the plasma membrane clusters, NHE1 is ubiquitous in the plasma membrane of almost all tissues, particularly the heart, the brain, and the peripheral nervous system [6]. Even though NHE1 is typically localized to the basolateral membrane of diverse epithelia, it is also found in the apical membrane of choroid plexus epithelia. In cardiomyocytes, NHE1 is located in intercalated disks and T-tubules rather than in the peripheral sarcolemmal membranes, which may influence local pH [3].
1.2. NHE1 Structure
A detailed insight into the structure of NHE1 is imperative for understanding its mechanisms and developing more powerful inhibitors. Nevertheless, the high-resolution structure of NHE1 remains elusive due to difficulties in crystallizing the protein molecules. The primary structure of human NHE1 was first described by Claude Sardet et al. They predicted that human NHE1 had 815 amino acids, with the first 500 residues comprising 10 transmembranes (TMs) helixes, and the hydrophilic C-terminal tail of 315 residues comprising a regulatory domain, based on hydropathy analysis [7]. Shrode et al. [8] confirmed the intracellular location of the NH2 and COOH termini via chymotryptic cleavage and epitope tagging. Moreover, the region between TM9–10 and TM11–12 was not completely available from the outside of the cells, suggesting that it might be buried inside the membrane. Based on experimentation using substituted cysteine accessibility examination, Wakabayashi et al. [9] proposed a novel model for the topology of NHE1 with the N- and C-termini situated in the cytol and the N-glycosylation site at N75. Na+ and H+ exchange is carried out by 12 relatively conserved TM segments joined by short loops and two long re-entrant loops, while 315 residues compose a more divergent hydrophilic C-terminal cytosolic domain that plays a regulatory role, and 15 residues form an extremely short hydrophobic N-terminal tail that extends into the cell [2]. Additionally, there are several binding sites in the C-terminal domain, such as esrin/radixin/moesin (ERM), calmodulin (CaM), calcineurin (CaN), and calcineurin homologous protein (CHP) [10]. The model of NHE1 is shown in Figure 1. The greatest potential breakthrough stems from elucidating the high-resolution structures of the bacterial Na+/H+ antiporters, including the well-known antiporter Escherichia coli (Ec)NhaA, which are easy to overproduce and crystallize. The bacterial EcNhaA and eukaryotic Na+/H+ exchanger play similar roles in the regulation of pHi and electrolyte homeostasis and thus are considered to share a similar structural folding [4][11]. Later, Landau et al. developed a novel topological model of NHE1 [12]. It was obtained computationally, including a revolutionary conserved analysis and a folding comparison with NhaA [13], comparable to Wakabayashi’s model with 12 TM domains, although amino acids 1-125 were considered to be eliminated by cleavage. In this model, the TM9 (amino acid 341-362) of Wakabayashi’s model was redistributed into two helices, TM7 and TM8, and the re-entrant segment extracellular loop (EL) 5 was reassigned to TM9. The last three TMs are identical in these two models [11]. Liu et al. [14] used cysteine scanning accessibility and detection of glycosylation of mature proteins to characterize the NHE1 protein, and they demonstrated that the basic features of Wakabayashi were correct. The latest three-dimensional model of human NHE1 was predicted based on the known structure of Methanocaldoccocus jannaschii (more similar in sequence and function to mammalian SLC9As than EcNhA) and combined with biochemical surface accessible data [15]. The specific model description was reviewed in Dutta’s [11].
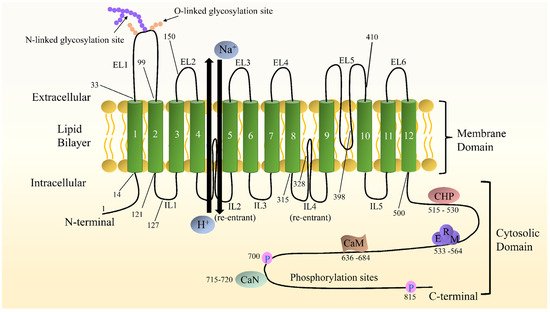
Figure 1. Schematic diagram of NHE1 topological structure. Both the N- and C- termini are located in the cytoplasm. The numbers indicate predicted transmembrane domains. Some representative amino acids are pointed out. The C-terminal hydrophilic cytoplasmic structural domain contains binding sites for a variety of proteins, including calmodulin (CaM), calcineurin (CaN), calcineurin homologous protein (CHP), and esrin/radixin/moesin (ERM), as well as cytoplasmic structural domains involved in phosphorylation and activity regulation.
2. NHE1 in Cardiac Physiology Regulation
NHEs are the most extensively studied acid-base regulators in various mammalian cells, including cardiomyocytes. They act a pivotal part in regulating cytoplasmic acid-base homeostasis together with HCO3− transport systems, including Na+/HCO3− cotransporters (NBCs) and Cl−/HCO3− anion exchangers (AEs) [16]. In many cell types, NHE1 is the primary alkalinization mechanism, preventing the destructive effects of excessive acidification. The intracellular acid load generated by normal cardiomyocyte metabolism activates NHE1 protein to transport one intracellular proton inside the cells and one extracellular Na+ outside the cells, thereby preventing cellular acidosis. In this process, the inward Na+ gradient created by the Na+/K+ ATPase drives the counter transport of proton from the cytoplasm [17]. Furthermore, NHE1 is also engaged in regulating cardiomyocyte volume, growth, proliferation, apoptosis, and differentiation, as well as a series of physiological activities.
Cell volume is an important component of an organism’s homeostasis. Volume regulation is crucial to cardiomyocyte function in healthy and disease states. Fundamental metabolic processes, such as respiration, constantly produce osmotic active products, which, if not regulated, would flow into the cell, leading to its swelling [18]. In addition, various environmental conditions can contribute to alterations in intracellular and intravascular volume. Swelling or shrinkage of cells may weaken the integrity of the cell membrane. It is hypothesized that the shrinkage of cells in response to a hyperosmotic extracellular milieu may be counterbalanced by an increased influx of Na+ and Cl−, accompanied by the osmolarity-driven water influx and compensatory swelling. Known as regulatory volume increase (RVI), this process serves to counteract the decrease of cell volume [2][18]. Other transporters have also been involved in RVI, including Na+/K+/2Cl− cotransporters (NKCC), electroneutral anion exchanger Cl−/HCO3− (AE2) [2]. The H+ excretion through NHEs and HCO3− exiting through AE2 are supplemented in the cell by H2CO3, which is easily produced from CO2, the product of aerobic metabolism. This process causes the entry of NaCl. On the other hand, Na+ entering the cells through NKCC and NHEs is pumped out by Na+/K+ ATPase to exchange K+, which ultimately results in the uptake of KCl by the cells [10]. In addition, the NKCC isoforms NKCC1 and NKCC2 and NHE isoforms NHE1, 2, and 4 are shown to be contraction-activated, whereas NHE3 and 5 are manifested as contraction-inhibited [17][19]. It has been demonstrated that NHE1 activation caused by cell contraction depends on the activity of CaM, which is likely downstream of tyrosine kinase Janus kinase 2(JAK2) and requires active phospholipase C (PLC).
The progression of the cell cycle is hypersensitive to cytoplasmic pH and is eliminated at acidic pH [20]. As a result, NHE1 has been widely shown to play a role in cell cycle progression and cell proliferation. The role of NHE1 in cell proliferation was originally theorized based on its growth-promoting effect and was reckoned to be related to the mitogen-induced increase of pHi. It has been found that the role of NHE1 in ion translocation is crucial for conferring enhancement of proliferation [21]. The proliferation of NHE1-deficient cells is severely diminished, and the G2-M checkpoint is delayed [22]. However, it is not completely understood how NHE1 regulates cell proliferation. It has been demonstrated that an increase in pHi may promote cell proliferation by facilitating protein synthesis. Concerning the role of cell volume, increases in cell volume and the uptake of inorganic ions are partially required for cell division, and this effect may be mediated by NHE1 [23]. NHE1 may impact cell proliferation by enhancing cell survival or suppressing apoptosis. Cell contraction and intracellular acidification are indications of apoptosis, whereas NHE1, increased regulatory volume, and intracellular alkalization may serve as an antiapoptotic signal [21]. The decrease in cytoplasmic volume results in NHE1-regulated Na+/H+ exchange, leading to phosphorylation and recruitment of ERM linkage to the cytosolic tail of NHE1. The interaction of ERM-NHE1 results in the development of a signaling complex, such as Akt and phosphatidylinositol 3-kinase (PI3K) which phosphorylates various substrates, contributing to the inhibition of apoptosis [24].
An intriguing area of research underway questions the processes of cell differentiation, some of which indicate the involvement of NHE1. Inhibition or deletion of NHE1 could impair the differentiation pathway [25]. It has been demonstrated that NHE1 activity promotes the differentiation of stem cells into the cardiomyocyte lineages. Elevated expression of NHE1 seems to have a role in promoting cardiomyocyte development [26]. NHE1 appears to either promote or prevent programmed cell death, depending on the cell type [27]. Furthermore, NHE1 has been assigned central roles in cytoskeletal tissue and cell motility. The cytoplasmic tail of NHE1 binds to ERM proteins and acts as an anchor for actin filaments. When these connections are disrupted or NHE1 activity is inhibited, external adhesion formation and cell migration are inhibited [22].
References
- Mithell, P.; Moyle, J. Acid-Base titration across the membrane system of rat-liver mitochondria. Biochem. J. 1967, 104, 588–600.
- Parker, M.D.; Myers, E.J.; Schelling, J.R. Na+-H+ exchanger-1 (NHE1) regulation in kidney proximal tubule. Cell Mol. Life Sci. 2015, 72, 2061–2074.
- Donowitz, M.; Ming Tse, C.; Fuster, D. SLC9/NHE gene family, a plasma membrane and organellar family of Na+/H+ exchangers. Mol. Asp. Med. 2013, 34, 236–251.
- Hendus-Altenburger, R.; Kragelund, B.B.; Pedersen, S.F. Structural dynamics and regulation of the mammalian SLC9A family of Na+/H+ exchangers. Curr. Top. Membr. 2014, 73, 69–148.
- Lee, S.H.; Kim, T.; Park, E.S.; Yang, S.; Jeong, D.; Choi, Y.; Rho, J. NHE10, an osteoclast-specific member of the Na+/H+ exchanger family, regulates osteoclast differentiation and survival. Biochem. Biophys. Res. Commun. 2008, 369, 320–326.
- Fliegel, L. Structural and functional changes in the Na+/H+ exchanger isoform 1, induced by Erk1/2 phosphorylation. Int. J. Mol. Sci. 2019, 20, 2378.
- Sardet, C.; Franchi, A.; Pouysségur, J. Molecular cloning, primary structure, and expression of the human growth factor-activatable Na+/H+ antiporter. Cell 1989, 56, 271–280.
- Shrode, L.D.; Gan, B.S.; D’Souza, S.J.; Orlowski, J.; Grinstein, S. Topological analysis of NHE1, the ubiquitous Na+/H+ exchanger using chymotryptic cleavage. Am. J. Physiol. Cell Physiol. 1998, 275, C431–C439.
- Wakabayashi, S.; Pang, T.; Su, X.; Shigekawa, M. A novel topology model of the human Na+/H+ exchanger isoform 1. J. Biol. Chem. 2000, 275, 7942–7949.
- Li, T.; Tuo, B. Pathophysiology of hepatic Na+/H+ exchange (Review). Exp. Ther. Med. 2020, 20, 1220–1229.
- Dutta, D.; Fliegel, L. Molecular modeling and inhibitor docking analysis of the Na+/H+ exchanger isoform one. Biochem. Cell Biol. 2019, 97, 333–343.
- Landau, M.; Herz, K.; Padan, E.; Ben-Tal, N. Model structure of the Na+/H+ exchanger 1 (NHE1): Functional and clinical implications. J. Biol. Chem. 2007, 282, 37854–37863.
- Lee, B.L.; Sykes, B.D.; Fliegel, L. Structural analysis of the Na+/H+ exchanger isoform 1 (NHE1) using the divide and conquer approach. Biochem. Cell Biol. 2011, 89, 189–199.
- Liu, Y.; Basu, A.; Li, X.; Fliegel, L. Topological analysis of the Na+/H+ exchanger. Biochim. Biophys. Acta 2015, 1848, 2385–2393.
- Fliegel, L. Role of genetic mutations of the Na+/H+ exchanger isoform 1, in human isease and protein targeting and activity. Mol. Cell Biochem. 2021, 476, 1221–1232.
- Orlowski, J.; Grinstein, S. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflug. Arch. 2004, 447, 549–565.
- Orlowski, J.; Grinstein, S. Na+/H+ exchangers. Compr. Physiol. 2011, 1, 2083–2100.
- Alexander, R.T.; Grinstein, S. Na+/H+ exchangers and the regulation of volume. Acta Physiol. 2006, 187, 159–167.
- Hoffmann, E.K.; Pedersen, S.F. Cell volume homeostatic mechanisms: Effectors and signalling pathways. Acta Physiol. 2011, 202, 465–485.
- Putney, L.K.; Barber, D.L. Expression profile of genes regulated by activity of the Na-H exchanger NHE1. BMC Genom. 2004, 5, 46.
- Putney, L.K.; Denker, S.P.; Barber, D.L. The changing face of the Na+/H+ exchanger, NHE1: Structure, regulation, and cell actions. Annu. Rev. Pharmacol. Toxicol. 1999, 42, 527–552.
- Fliegel, L. The Na+/H+ exchanger isoform 1. Int. J. Biochem. Cell Biol. 2005, 37, 33–37.
- Pedersen, S.F. The Na+/H+ exchanger NHE1 in stress-induced signal transduction: Implications for cell proliferation and cell death. Pflug. Arch. 2006, 452, 249–259.
- Schelling, J.R.; Abu Jawdeh, B.G. Regulation of cell survival by Na+/H+ exchanger-1. Am. J. Physiol. Ren. Physiol. 2008, 295, F625–F632.
- Pedersen, S.F.; Counillon, L. The SLC9A-C mammalian Na+/H+ exchanger family: Molecules, mechanisms, and physiology. Physiol. Rev. 2019, 99, 2015–2113.
- Li, X.; Karki, P.; Lei, L.; Wang, H.; Fliegel, L. Na+/H+ exchanger isoform 1 facilitates cardiomyocyte embryonic stem cell differentiation. Am. J. Physiol. Heart Circ. Physiol. 2009, 296, H159–H170.
- Malo, M.E.; Fliegel, L. Physiological role and regulation of the Na+/H+ exchanger. Can. J. Physiol. Pharmacol. 2006, 84, 1081–1095.
More
