Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Catherine Yang and Version 1 by Nan-Shan Chang.
Tumor suppressor WWOX inhibits cancer growth and retards Alzheimer’s disease (AD) progression. Supporting evidence shows that the more strongly WWOX binds intracellular protein partners, the weaker is cancer cell growth in vivo. Whether this correlates with retardation of AD progression is unknown. Two functional forms of WWOX exhibit opposite functions. pY33-WWOX is proapoptotic and anticancer, and is essential for maintaining normal physiology.
- tumor suppressor
- p53
- WWOX
1. WW Domain-Containing Oxidoreductase (WWOX)
WWOX and its binding proteins form a complicated signaling network which is needed for normal physiology and/or disease progression [11,12,13,14,15,16,17,18,19,20,21][1][2][3][4][5][6][7][8][9][10][11]. WWOX was first discovered in 2000 [12][2]. Most recently, the human WWOX gene has been defined as a risk factor for AD [22][12]. Currently, there are many outstanding review articles documented in the PubMed database. In brief, WWOX protein possesses two N-terminal WW domains, a C-terminal short chain alcohol dehydrogenase/reductase (SDR) domain, and a nuclear localization signal present in between the WW domains [12][2] (Figure 21A). A mitochondria-targeting region is located in the SDR domain. Both WW and SDR domains have their own specific protein-binding partners, as reviewed in a recent article [19][9]. The first WW domain (WW1) possesses two tryptophans and binds PPXY (e.g., PPPY) or LPXY motif [12,23][2][13] motif in the target proteins, where X is any amino acid. However, when Y33 in the WW1 becomes phosphorylated, pY33-WWOX has an expanded binding capability with numerous proteins.
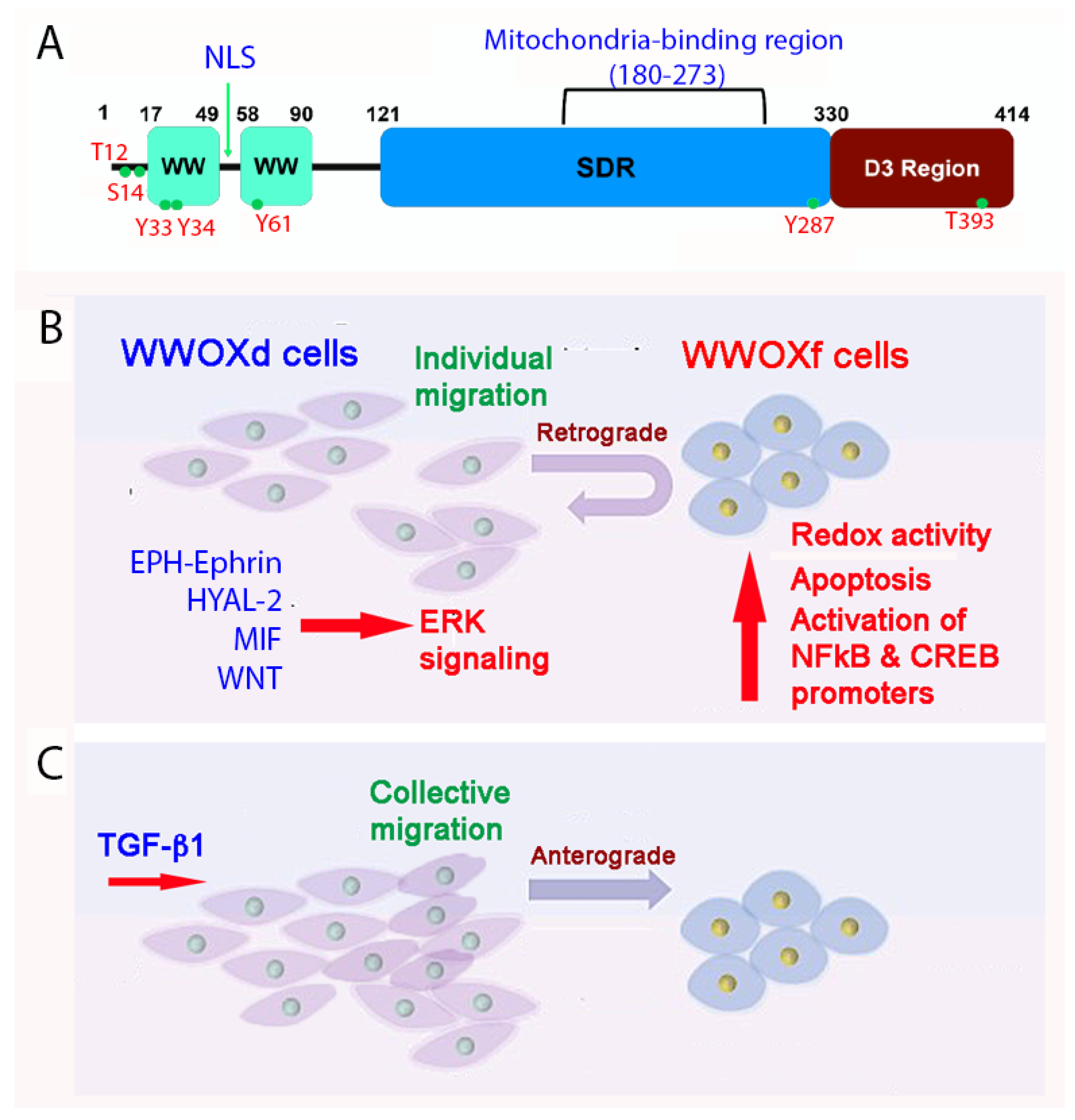
Figure 21. A schematic model for how WWOXd cells undergo retrograde migration upon facing WWOXf cells. (A) The primary structure of WWOX is depicted. (B) When WWOXd cells face WWOXf cells, WWOXd cells undergo activation of multiple signaling pathways, including MIF, HYAL-2, WNT and EPH-Ephrin, that converge to MEK/ERK pathway leading to retrograde migration. WWOXd cells have no physical contacts with WWOXf cells but are able to kill WWOXf cells from a distance. (C) TGF-β abolishes the retrograde migration and apoptosis, and induces anterograde migration and merger of WWOXd and WWOXf cells. Similarly, antibodies against MIF, HYAL-2 or EPH-Ephrin, and WNT inhibitors abolish the retrograde migration of WWOXd cells in facing WWOXf cells [36,37][14][15].
Overall, WWOX participates in many ligand/receptor-mediated signaling pathways such as WNT/β-catenin [24][16], JNK [25][17], TGFβ1/HYAL-2 [26][18], TGFβ/SMADs [26,27][18][19], HIPPO [28][20], HCG/MET [29][21], ], JAK2/STAT3 [30[22][23][24],31,32], p73 [33][25], and others. WW2 domain has only one tryptophan and its function is largely unknown. WW2 is proposed to line up with WW1 in facilitating the binding of WWOX with its interacting partners [34][26]. The SDR domain has an NSYK (Asn-Ser-Tyr-Lys) motif, which binds sex steroid hormones androgen and estrogen [12][2]. The SDR domain also exhibits redox activity [35[14][15][27],36,37], and binds tau and GSK-3β to prevent tau hyperphosphorylation [19,38,39,40][9][28][29][30]. Whether the redox activity of SDR domain limits GSK-3β-mediated tau hyperphosphorylation and subsequent tau aggregation is unknown [38,40][28][30].
2. WWOX Controls Cell Migration, Cell-Cell Recognition, and Neuronal Heterotopia
Of utmost concern is that human newborns with WWOX gene deficiency suffer severe neural diseases such as seizure, encephalopathy, and early death [19,20,21,41,42][9][10][11][31][32]. There is no cure for the disease. Loss of WWOX enhances cell mobility [30,36][14][22]. WWOX gene deficiency in newborns results in neuronal migration disorders (or neuronal heterotopia) that lead to epileptic seizures [40,42][30][32]. Two types of cells based upon their expression of functional or dysfunctional WWOX have recently been identified [36,38][14][28]. Cells which are deficient in WWOX protein, or express dysfunctional WWOX, can be considered as metastatic cancer cells (designated as WWOXd).
At room temperature, WWOXd cells are less efficient in generating Ca2+ influx and undergo non-apoptotic explosion in response to UV irradiation. In contrast, functional WWOX-expressing cells (designated WWOXf) exhibit non-apoptotic nucleus-dependent bubbling cell death (BCD) [17][7] and efficient Ca2+ influx caused by UV or apoptotic stress at room temperature [36][14]. Formation of a nitric oxide (NO)-containing nuclear bubble per cell during BCD is due to UV-induced upregulation of NO synthase 2 (NOS2) [17][7]. WWOXf cells, which are mainly normal and benign cancer cells, migrate collectively and force the individually migrating WWOXd cells to undergo retrograde migration [36,37] (Figure 2B,C)[14][15]. WWOXd cells, in return, induce WWOXf cells to undergo apoptosis from a distance without physical contact. The cytokines responsible for the induced apoptosis are unknown [36,37][14][15]. During cell-to-cell encounter, WWOXd cells exhibit activation of MIF, HYAL-2, Eph, and Wnt pathways, which converge to the MEK/ERK signaling and enables the cells to move away from WWOXf cells (Figure 2B,C). Specific antibodies against MIF or HYAL-2, or inhibitors for WNT, cause WWOXf to greet WWOXd cells, and both cells merge eventually (Figure 2B,C). Metastatic cancer cell-derived TGF-β1 allows merger of WWOXf with WWOXd cells [36,37][14][15]. A detailed signaling for the pathway of WNT, HYAL-2, EPH-Ephrin, EGF/EFGR, or MIF linking to ERK (or ERK1/2) is shown (Figure 32). The CD44/HYAL-2 complex prevents the binding of TGF-β1 with HYAL-2. CD44 does not appear to be involved in the HYAL-2/WWOX/SMAD4 signaling [18][8] (Figure 32). Together, these observations suggest WWOX controls cell migration and cell-to-cell recognition and plays a crucial role in neuronal heterotopia that contributes to epileptic seizure [30,35,36,37][14][15][22][27].
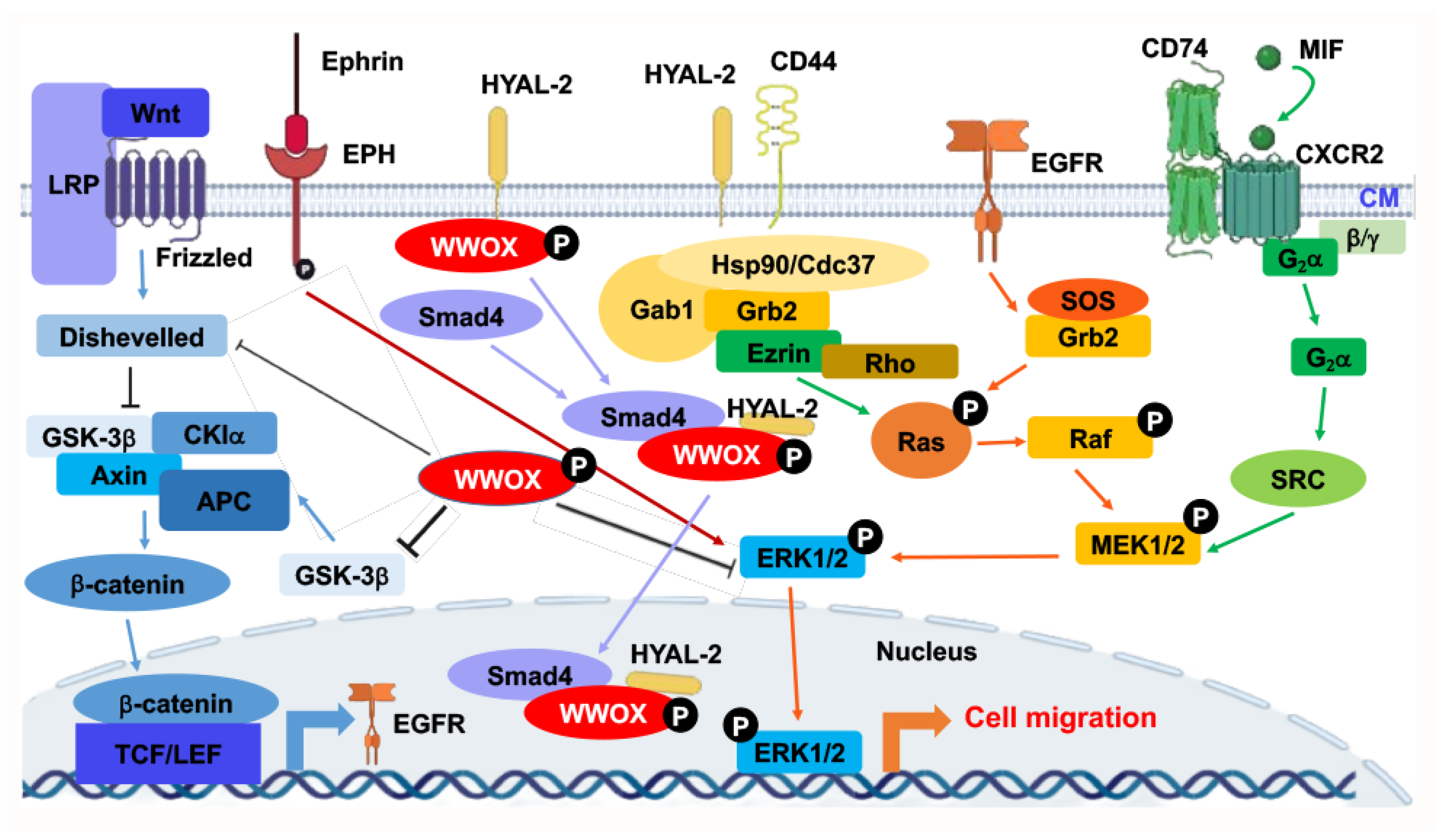
Figure 32. Signal pathways converged to ERK for controlling retrograde migration of WWOXd cells upon facing WWOXf cells. Pathways, including WNT, HYAL-2, Ephrin/EPH, EGF/EFGR, and MIF, allow signaling to drive down to ERK1/2. ERK1/2 controls the retrograde migration of WWOXd cells upon facing WWOXf cells. Suppression of WNT, MIF, HYAL-2, or Ephrin/EPH causes the retrograde migration of WWOXd cells, anterograde migration and finally merger with WWOXf cells [36,37][14][15]. The role of HYAL-2/CD44 in controlling retrograde migration remains to be established.
3. WWOX Signaling Network
By STRING analysis (https://string-db.org/cgi/network?taskId=brQHTnXWILzL&sessionId=b73ctYMesgwG) (accessed on 19 June 2022), the first level of WWOX-binding protein network reveals that WWOX has connection with 5 interactors, namely TP53, ERBB4, DVL2, FAM189B, and WBP2 [11,25,43,44,45,46,47,48][1][17][33][34][35][36][37][38]. The binding has been validated by experimental approaches (Figure 43). Further analysis expanding the interactors up to 26 and 136, respectively, shows many biological process and molecular functions, including beta-catenin destruction complex assembly, response to metformin, ezrin [49][39], and many others. Again, the network can be expanded tremendously. The significance of this approach is that the data show the high complexity in the biological network and yet precise generation of functional machinery. The downside is that the analysis does not show domain/domain-specific binding interactions. Binding affinity-based interaction network is not clear. The computational approach also fails to prove the empirical observations that the stronger the binding of WWOX with intracellular proteins, the better the suppression of cancer growth and retardation of AD progression [19][9].
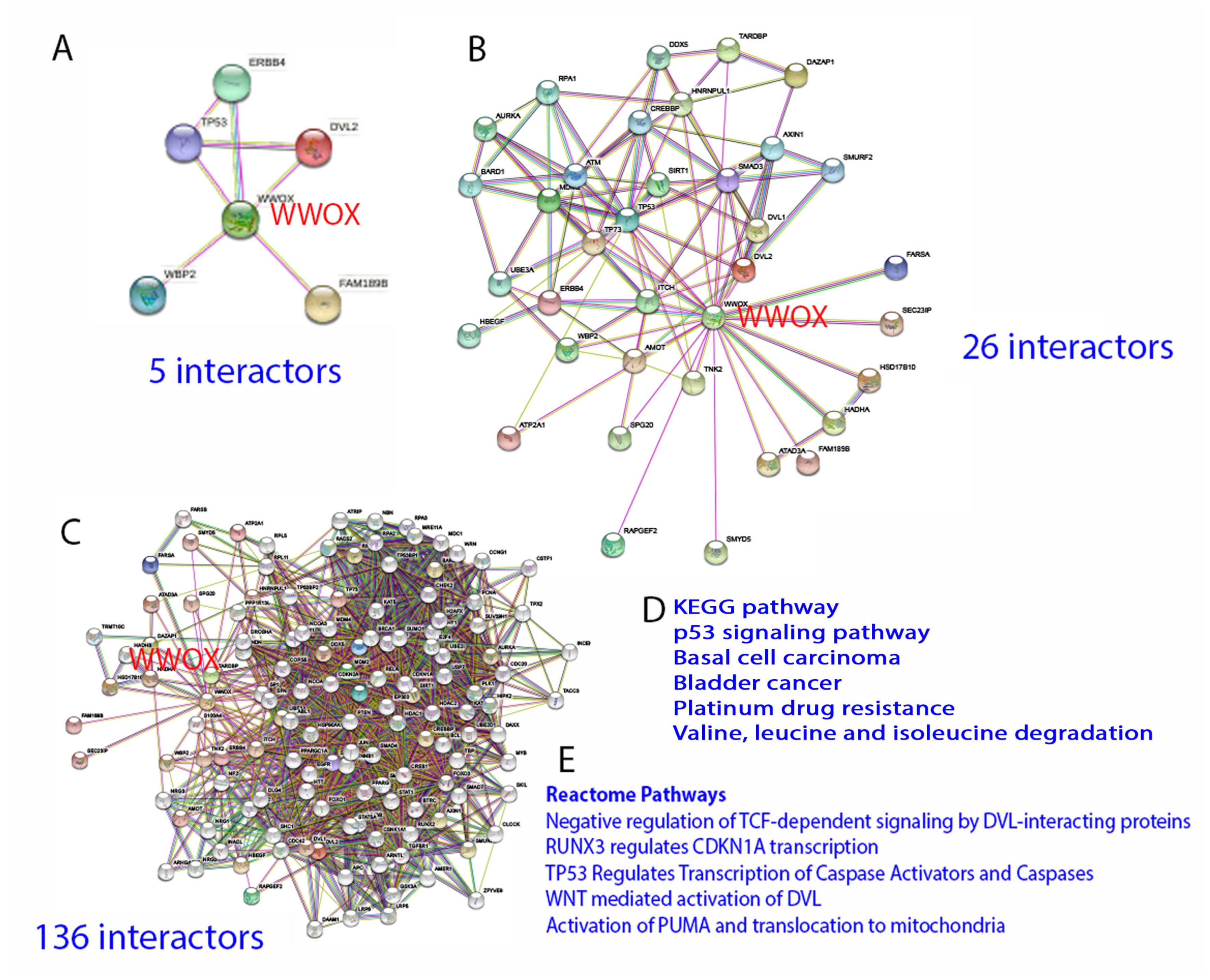
Figure 43. STRING analysis of WWOX signaling network. (A–E) The analysis starts with 5 interactors, and is then expanded to 26 and 136 interactors, respectively. Representative KEGG and Reactome pathways are shown.
4. WWOX Functional Measurement by Time-Lapse FRET Microscopy
Förster resonance energy transfer (FRET) microscopy is a feasible approach to measure WWOX function in a real-time manner. FRET can be utilized to determine spatial proximity among proteins either at a single or multiple protein levels in cultured cells in a real-time mode [18,43,44,55,56,57,58,59,60,61,62][8][33][34][40][41][42][43][44][45][46][47]. For example, binding of a CFP (cyan fluorescence protein)-tagged bait (or donor) protein with a YFP (yellow fluorescence protein)-tagged target (or acceptor) protein results in energy flow from the donor to the acceptor. In other words, an excitation wavelength is used to excite a donor protein (e.g., CFP tagged), and once excited the donor protein transfers the resulting energy to the acceptor (e.g., YFP tagged), which subsequently emits a longer wavelength with a lower energy. To be effective in signal transduction, protein proximity at 1–10 nm is needed for FRET detection. Furthermore, energy release from the first donor protein can excite two acceptors tagged with different fluorophores for determining parallel signaling pathways.
5. A WWOX7-21 Epitope Peptide Drives the HYAL-2/WWOX/SMAD4 Signaling
The mechanisms by which WWOX-mediated cancer suppression and inhibition of neurodegeneration take place are largely unknown. There are two surface exposed epitopes in WWOX, which are at amino acids #7-21 and #286-299 [36,53,54][14][48][49]. Synthetic WWOX7-21 peptide, or truncation down to 5-amino acid WWOX7-11, strongly blocks and prevents the growth and metastasis of melanoma and skin cancer cells in mice [54][49]. WWOX286-299 also inhibits cancer cell growth, whereas it fails to block cancer metastasis to the lung and liver [54][49]. By time-lapse microscopy, antibody against WWOX7-21 suppresses ceritinib-mediated breast 4T1 cell sphere explosion and death (Figure 64A,C). 4T1 cell spheres express many makers of stem cells (e.g., Sox2, Oct4 and Nanog). Ceritinib is an antineoplastic kinase inhibitor for treating anaplastic lymphoma kinase (ALK)-positive metastatic non-small cell lung cancer (NSCLC) [73][50]. In contrast, WWOX7-21 peptide potently enhances the function of ceritinib in causing the explosion and death of 4T1 cell spheres (Figure 64B,C) [54][49]. pS14-WWOX7-21 peptide or WWOX7-21 antibody supports cancer survival by blocking ceritinib cytotoxicity (Figure 64A–C). Further analysis reveals that ceritinib-mediated cell death is due to rapid upregulation of proapoptotic pY33-WWOX, downregulation of prosurvival pERK, prompt increases in Ca2+ influx, and disruption of the IκBα/WWOX/ERK prosurvival signaling [54][49]. WWOX7-11 (AGLDD) peptide does not block ceritinib cytotoxicity in vitro (Figure 64B). In stark contrast, WWOX7-11 is even more powerful than WWOX7-21 in blocking cancer cell growth in vivo [54][49]. The mechanism by which WWOX7-11 works in an opposite fashion in vitro and in vivo is not clear. Antibody against WWOX286-299 also enhances ceritinib-mediated 4T1 cell sphere explosion and death (Video S2). The observations indicate that WWOX with S14 phosphorylation is pro-survival for cancer cells [54][49].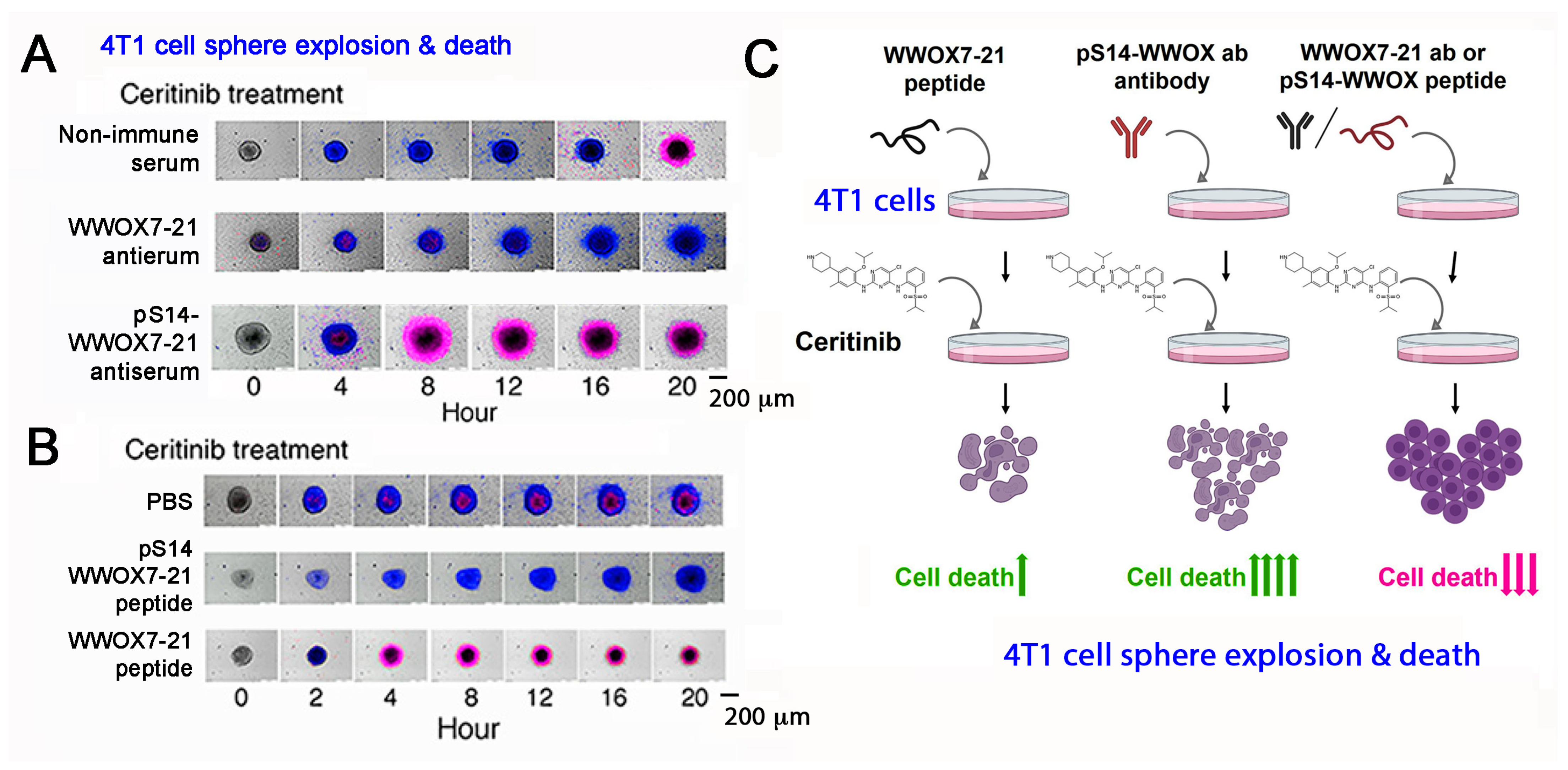
Figure 64. pS14-WWOX7-21 peptide and WWOX7-21 antibody block ceritinib-mediated breast 4T1 cell sphere explosion and death. Cells are pretreated with an indicated peptide (10 μM) or antiserum (1:100 dilution) for 30 min and then treated with ceritinib (30 μM) for time-lapse microscopy. Nuclear stains DAPI (blue) and Propidium Iodide (red) are added in the culture. (A,B) Both WWOX7-21 peptide and pS14-WWOX7-21 antibody enhance 4T1 cell sphere explosion and death. pS14-WWOX7-21 peptide strongly blocks ceritinib-mediated 4T1 sphere explosion and cell death. (C) Summary of enhancers and inhibitors for ceritinib-mediated cell death. Upward green arrows indicate increases in cell death, and downward red arrows for reduced cell death. (image data adapted from Reference [54][49].
To understand the functional mechanism for WWOX7-21-mediated cancer suppression, exogenous WWOX7-21 peptide colocalizes with membrane type II TGFβ receptor (TGFβRII) [54][49]. WWOX antibody or pY33-WWOX antibody pulls down TGFβRII, WWOX, and HYAL-2 in the lipid raft, indicating that TGFβRII is an additional component of the HYAL-2/WWOX/SMAD4 signaling [36][14]. WWOX7-21 peptide may undergo self-polymerization in vitro [54][49]. WWOX7-21 peptide binds the first WW domain of WWOX on the cell membrane [36][14]. Antibody against WWOX7-21 peptide pulls down the full-length WWOX, membrane HYAL-2 and TGFβRII, again further validating that WWOX7-21 peptide is able to initiate the HYAL-2/WWOX/SMAD4 signaling [36][14].
6. Phosphorylation Status of WWOX in the HYAL-2/WWOX/SMAD4 Complex and Disease Progression
Endogenous pS14-WWOX protein is accumulated in the lesions of growing tumors and AD brains [50,51][51][52]. Suppression of WWOX phosphorylation at S14 by Zfra4-10 peptide results in significant reduction in cancer growth in mice [51][52], and enhanced restoration of memory loss and mitigation of AD-like symptoms in triple transgenic (3xTg) mice [50][51]. pY33-WWOX, but not pS14-WWOX, is needed for binding with HYAL-2 and SMAD4 in vivo [36][14]. Whether pY33-WWOX is downregulated and pS14-WWOX upregulated in the HYAL-2/WWOX/SMAD4 complex during disease progression is unknown. Zfra, known as zinc finger-like protein that regulates apoptosis, is a 31-amino-acid protein [18,50,51,52,74,75][8][51][52][53][54][55]. As short as 7 amino acids, Zfra4-10 (RRSSSCK) strongly suppresses the progression of cancer and AD [50,51][51][52]. S8 is a determined phosphorylation site, and is a key to the function of Zfra [18,50,51][8][51][52]. Additionally, Zfra4-10 blocks NF-κB-mediated inflammation and accelerates degradation of proteins in the AD pathologies, as well as induction of the activation of HYAL-2+ CD3− CD19− lymphocytes to kill cancer in vivo [50,51,52,53][48][51][52][53].References
- Chang, N.S.; Pratt, N.; Heath, J.; Schultz, L.; Sleve, D.; Carey, G.B.; Zevotek, N. Hyaluronidase induction of a WW domain-containing oxidoreductase that enhances tumor necrosis factor cytotoxicity. J. Biol. Chem. 2001, 276, 3361–3370.
- Chang, N.S.; Hsu, L.J.; Lin, Y.S.; Lai, F.J.; Sheu, H.M. WW domain-containing oxidoreductase: A candidate tumor suppressor. Trends Mol. Med. 2007, 13, 12–22.
- Abu-Remaileh, M.; Joy-Dodson, E.; Schueler-Furman, O.; Aqeilan, R.I. Pleiotropic Functions of Tumor Suppressor WWOX in Normal and Cancer Cells. J. Biol. Chem. 2015, 290, 30728–30735.
- Hussain, T.; Lee, J.; Abba, M.C.; Chen, J.; Aldaz, C.M. Delineating WWOX Protein Interactome by Tandem Affinity Purification-Mass Spectrometry: Identification of Top Interactors and Key Metabolic Pathways Involved. Front. Oncol. 2018, 8, 591.
- Chang, H.T.; Liu, C.C.; Chen, S.T.; Yap, Y.V.; Chang, N.S.; Sze, C.I. WW domain-containing oxidoreductase in neuronal injury and neurological diseases. Oncotarget 2014, 5, 11792–11799.
- Chang, N.S. Bubbling cell death: A hot air balloon released from the nucleus in the cold. Exp. Biol. Med. 2016, 241, 1306–1315.
- Chen, S.J.; Lin, P.W.; Lin, H.P.; Huang, S.S.; Lai, F.J.; Sheu, H.M.; Hsu, L.J.; Chang, N.S. UV irradiation/cold shock-mediated apoptosis is switched to bubbling cell death at low temperatures. Oncotarget 2015, 6, 8007–8018.
- Hsu, L.J.; Chiang, M.F.; Sze, C.I.; Su, W.P.; Yap, Y.V.; Lee, I.T.; Kuo, H.L.; Chang, N.S. HYAL-2-WWOX-SMAD4 Signaling in Cell Death and Anticancer Response. Front. Cell Dev. Biol. 2016, 4, 141.
- Hsu, C.Y.; Lee, K.T.; Sun, T.Y.; Sze, C.I.; Huang, S.S.; Hsu, L.J.; Chang, N.S. WWOX and Its Binding Proteins in Neurodegeneration. Cells 2021, 10, 1781.
- Aldaz, C.M.; Hussain, T. WWOX Loss of Function in Neurodevelopmental and Neurodegenerative Disorders. Int. J. Mol. Sci. 2020, 21, 8922.
- Liu, C.C.; Ho, P.C.; Lee, I.T.; Chen, Y.A.; Chu, C.H.; Teng, C.C.; Wu, S.N.; Sze, C.I.; Chiang, M.F.; Chang, N.S. WWOX Phosphorylation, Signaling, and Role in Neurodegeneration. Front. Neurosci. 2018, 12, 563.
- Kunkle, B.W.; Grenier-Boley, B.; Sims, R.; Bis, J.C.; Damotte, V.; Naj, A.C.; Boland, A.; Vronskaya, M.; van der Lee, S.J.; Amlie-Wolf, A.; et al. Genetic meta-analysis of diagnosed Alzheimer’s disease identifies new risk loci and implicates Aβ, tau, immunity and lipid processing. Nat. Genet. 2019, 51, 414–430.
- Abu-Odeh, M.; Bar-Mag, T.; Huang, H.; Kim, T.; Salah, Z.; Abdeen, S.K.; Sudol, M.; Reichmann, D.; Sidhu, S.; Kim, P.M.; et al. Characterizing WW domain interactions of tumor suppressor WWOX reveals its association with multiprotein networks. J. Biol. Chem. 2014, 289, 8865–8880.
- Chen, Y.A.; Sie, Y.D.; Liu, T.Y.; Kuo, H.L.; Chou, P.Y.; Chen, Y.J.; Lee, K.T.; Chen, P.J.; Chen, S.T.; Chang, N.S. Normal cells repel WWOX-negative or -dysfunctional cancer cells via WWOX cell surface epitope 286–299. Commun. Biol. 2021, 4, 753.
- Chou, P.Y.; Lai, F.J.; Chen, Y.A.; Sie, Y.D.; Kuo, H.L.; Su, W.P.; Wu, C.Y.; Liu, T.Y.; Wen, K.Y.; Hsu, L.J.; et al. Strategies by which WWOX-deficient metastatic cancer cells utilize to survive via dodging, compromising, and causing damage to WWOX-positive normal microenvironment. Cell Death Discov. 2019, 5, 97.
- Bouteille, N.; Driouch, K.; Hage, P.E.; Sin, S.; Formstecher, E.; Camonis, J.; Lidereau, R.; Lallemand, F. Inhibition of the Wnt/beta-catenin pathway by the WWOX tumor suppressor protein. Oncogene 2009, 28, 2569–2580.
- Chang, N.S.; Doherty, J.; Ensign, A. JNK1 physically interacts with WW domain-containing oxidoreductase (WOX1) and inhibits WOX1-mediated apoptosis. J. Biol. Chem. 2003, 278, 9195–9202.
- Hsu, L.J.; Schultz, L.; Hong, Q.; Van Moer, K.; Heath, J.; Li, M.Y.; Lai, F.J.; Lin, S.R.; Lee, M.H.; Lo, C.P.; et al. Transforming growth factor beta1 signaling via interaction with cell surface Hyal-2 and recruitment of WWOX/WOX1. J. Biol. Chem. 2009, 284, 16049–16059.
- Aldaz, C.M.; Ferguson, B.W.; Abba, M.C. WWOX at the crossroads of cancer, metabolic syndrome related traits and CNS pathologies. Biochim. Biophys. Acta 2014, 1846, 188–200.
- Chen, Y.A.; Lu, C.Y.; Cheng, T.Y.; Pan, S.H.; Chen, H.F.; Chang, N.S. WW Domain-Containing Proteins YAP and TAZ in the Hippo Pathway as Key Regulators in Stemness Maintenance, Tissue Homeostasis, and Tumorigenesis. Front. Oncol. 2019, 9, 60.
- Bendinelli, P.; Maroni, P.; Matteucci, E.; Desiderio, M.A. HGF and TGFβ1 differently influenced Wwox regulatory function on Twist program for mesenchymal-epithelial transition in bone metastatic versus parental breast carcinoma cells. Mol. Cancer 2015, 14, 112.
- Chang, R.; Song, L.; Xu, Y.; Wu, Y.; Dai, C.; Wang, X.; Sun, X.; Hou, Y.; Li, W.; Zhan, X.; et al. Loss of Wwox drives metastasis in triple-negative breast cancer by JAK2/STAT3 axis. Nat. Commun. 2018, 9, 3486.
- Taouis, K.; Driouch, K.; Lidereau, R.; Lallemand, F. Molecular Functions of WWOX Potentially Involved in Cancer Development. Cells 2021, 10, 1051.
- Abu-Odeh, M.; Salah, Z.; Herbel, C.; Hofmann, T.G.; Aqeilan, R.I. WWOX, the common fragile site FRA16D gene product, regulates ATM activation and the DNA damage response. Proc. Natl. Acad. Sci. USA 2014, 111, 4716–4725.
- Aqeilan, R.I.; Pekarsky, Y.; Herrero, J.J.; Palamarchuk, A.; Letofsky, J.; Druck, T.; Trapasso, F.; Han, S.Y.; Melino, G.; Huebner, K.; et al. Functional association between Wwox tumor suppressor protein and p73, a p53 homolog. Proc. Natl. Acad. Sci. USA 2004, 101, 4401–4406.
- Schuchardt, B.J.; Mikles, D.C.; Bhat, V.; McDonald, C.B.; Sudol, M.; Farooq, A. Allostery mediates ligand binding to WWOX tumor suppressor via a conformational switch. J. Mol. Recognit. 2015, 28, 220–231.
- Chou, P.Y.; Lin, S.R.; Lee, M.H.; Schultz, L.; Sze, C.I.; Chang, N.S. A p53/TIAF1/WWOX triad exerts cancer suppression but may cause brain protein aggregation due to p53/WWOX functional antagonism. Cell Commun. Signal. 2019, 17, 76.
- Sze, C.I.; Su, M.; Pugazhenthi, S.; Jambal, P.; Hsu, L.J.; Heath, J.; Schultz, L.; Chang, N.S. Down-regulation of WW domain-containing oxidoreductase induces Tau phosphorylation in vitro. A potential role in Alzheimer’s disease. J. Biol. Chem. 2004, 279, 30498–30506.
- Wang, H.Y.; Juo, L.I.; Lin, Y.T.; Hsiao, M.; Lin, J.T.; Tsai, C.H.; Tzeng, Y.H.; Chuang, Y.C.; Chang, N.S.; Yang, C.N.; et al. WW domain-containing oxidoreductase promotes neuronal differentiation via negative regulation of glycogen synthase kinase 3β. Cell Death Differ. 2012, 19, 1049–1059.
- Cheng, Y.Y.; Chou, Y.T.; Lai, F.J.; Jan, M.S.; Chang, T.H.; Jou, I.M.; Chen, P.S.; Lo, J.Y.; Huang, S.S.; Chang, N.S.; et al. Wwox deficiency leads to neurodevelopmental and degenerative neuropathies and glycogen synthase kinase 3β-mediated epileptic seizure activity in mice. Acta Neuropathol. Commun. 2020, 8, 6.
- Steinberg, D.J.; Aqeilan, R.I. WWOX-Related Neurodevelopmental Disorders: Models and Future Perspectives. Cells 2021, 10, 3082.
- Repudi, S.; Steinberg, D.J.; Elazar, N.; Breton, V.L.; Aquilino, M.S.; Saleem, A.; Abu-Swai, S.; Vainshtein, A.; Eshed-Eisenbach, Y.; Vijayaragavan, B.; et al. Neuronal deletion of Wwox, associated with WOREE syndrome, causes epilepsy and myelin defects. Brain 2021, 144, 3061–3077.
- Li, M.Y.; Lai, F.J.; Hsu, L.J.; Lo, C.P.; Cheng, C.L.; Lin, S.R.; Lee, M.H.; Chang, J.Y.; Subhan, D.; Tsai, M.S.; et al. Dramatic co-activation of WWOX/WOX1 with CREB and NF-kappaB in delayed loss of small dorsal root ganglion neurons upon sciatic nerve transection in rats. PLoS ONE 2009, 4, e7820.
- Hsu, L.J.; Hong, Q.; Chen, S.T.; Kuo, H.L.; Schultz, L.; Heath, J.; Lin, S.R.; Lee, M.H.; Li, D.Z.; Li, Z.L.; et al. Hyaluronan activates Hyal-2/WWOX/Smad4 signaling and causes bubbling cell death when the signaling complex is overexpressed. Oncotarget 2017, 8, 19137–19155.
- Aqeilan, R.I.; Donati, V.; Palamarchuk, A.; Trapasso, F.; Kaou, M.; Pekarsky, Y.; Sudol, M.; Croce, C.M. WW domain-containing proteins, WWOX and YAP, compete for interaction with ErbB-4 and modulate its transcriptional function. Cancer Res. 2005, 65, 6764–6772.
- Yang, T.; Xu, R.; Huo, J.; Wang, B.; Du, X.; Dai, B.; Zhu, M.; Zhan, Y.; Zhang, D.; Zhang, Y. WWOX activation by toosendanin suppresses hepatocellular carcinoma metastasis through JAK2/Stat3 and Wnt/β-catenin signaling. Cancer Lett. 2021, 513, 50–62.
- Zhang, H.; Huang, C.J.; Tian, Y.; Wang, Y.P.; Han, Z.G.; Li, X.C. Ectopic overexpression of COTE1 promotes cellular invasion of hepatocellular carcinoma. Asian Pac. J. Cancer Prev. 2012, 13, 5799–5804.
- Chang, N.S.; Doherty, J.; Ensign, A.; Schultz, L.; Hsu, L.J.; Hong, Q. WOX1 is essential for tumor necrosis factor-, UV light-, staurosporine-, and p53-mediated cell death, and its tyrosine 33-phosphorylated form binds and stabilizes serine 46-phosphorylated p53. J. Biol. Chem. 2005, 280, 43100–43108.
- Jin, C.; Ge, L.; Ding, X.; Chen, Y.; Zhu, H.; Ward, T.; Wu, F.; Cao, X.; Wang, Q.; Yao, X. PKA-mediated protein phosphorylation regulates ezrin-WWOX interaction. Biochem. Biophys. Res. Commun. 2006, 341, 784–791.
- Cheng, P.C. The Contrast Formation in Optical Microscopy. In Handbook of Biological Confocal Microscopy; Pawley, J.B., Ed.; Springer: Boston, MA, USA, 2006; pp. 162–206.
- Huang, S.S.; Su, W.P.; Lin, H.P.; Kuo, H.L.; Wei, H.L.; Chang, N.S. Role of WW Domain-containing Oxidoreductase WWOX in Driving T Cell Acute Lymphoblastic Leukemia Maturation. J. Biol. Chem. 2016, 291, 17319–17331.
- Che, Y.; Khavari, P.A. Research Techniques Made Simple: Emerging Methods to Elucidate Protein Interactions through Spatial Proximity. J. Investig. Dermatol. 2017, 137, e197–e203.
- Kuo, H.L.; Ho, P.C.; Huang, S.S.; Chang, N.S. Chasing the signaling run by tri-molecular time-lapse FRET microscopy. Cell Death Discov. 2018, 4, 45.
- Liu, Q.; Zheng, J.; Sun, W.; Huo, Y.; Zhang, L.; Hao, P.; Wang, H.; Zhuang, M. A proximity-tagging system to identify membrane protein-protein interactions. Nat. Methods 2018, 15, 715–722.
- Imani, M.; Mohajeri, N.; Rastegar, M.; Zarghami, N. Recent advances in FRET-Based biosensors for biomedical applications. Anal. Biochem. 2021, 630, 114323.
- Feng, X.A.; Poyton, M.F.; Ha, T. Multicolor single-molecule FRET for DNA and RNA processes. Curr. Opin. Struct. Biol. 2021, 70, 26–33.
- Nakamura, A.; Goto, Y.; Kondo, Y.; Aoki, K. Shedding light on developmental ERK signaling with genetically encoded biosensors. Development 2021, 148, dev199767.
- Su, W.P.; Wang, W.J.; Chang, J.Y.; Ho, P.C.; Liu, T.Y.; Wen, K.Y.; Kuo, H.L.; Chen, Y.J.; Huang, S.S.; Subhan, D.; et al. Therapeutic Zfra4-10 or WWOX7-21 Peptide Induces Complex Formation of WWOX with Selective Protein Targets in Organs that Leads to Cancer Suppression and Spleen Cytotoxic Memory Z Cell Activation In Vivo. Cancers 2020, 12, 2189.
- Wang, W.J.; Ho, P.C.; Nagarajan, G.; Chen, Y.A.; Kuo, H.L.; Subhan, D.; Su, W.P.; Chang, J.Y.; Lu, C.Y.; Chang, K.T.; et al. WWOX Possesses N-Terminal Cell Surface-Exposed Epitopes WWOX(7-21) and WWOX(7-11) for Signaling Cancer Growth Suppression and Prevention In Vivo. Cancers 2019, 11, 1818.
- Deeks, E.D. Ceritinib: A Review in ALK-Positive Advanced NSCLC. Target. Oncol. 2016, 11, 693–700.
- Lee, M.H.; Shih, Y.H.; Lin, S.R.; Chang, J.Y.; Lin, Y.H.; Sze, C.I.; Kuo, Y.M.; Chang, N.S. Zfra restores memory deficits in Alzheimer’s disease triple-transgenic mice by blocking aggregation of TRAPPC6AΔ, SH3GLB2, tau, and amyloid β, and inflammatory NF-κB activation. Alzheimers Dement. 2017, 3, 189–204.
- Lee, M.H.; Su, W.P.; Wang, W.J.; Lin, S.R.; Lu, C.Y.; Chen, Y.A.; Chang, J.Y.; Huang, S.S.; Chou, P.Y.; Ye, S.R.; et al. Zfra activates memory Hyal-2+ CD3- CD19- spleen cells to block cancer growth, stemness, and metastasis in vivo. Oncotarget 2015, 6, 3737–3751.
- Su, W.P.; Wang, W.J.; Sze, C.I.; Chang, N.S. Zfra induction of memory anticancer response via a novel immune cell. Oncoimmunology 2016, 5, e1213935.
- Hsu, L.J.; Schultz, L.; Mattison, J.; Lin, Y.S.; Chang, N.S. Cloning and characterization of a small-size peptide Zfra that regulates the cytotoxic function of tumor necrosis factor by interacting with JNK1. Biochem. Biophys. Res. Commun. 2005, 327, 415–423.
- Hsu, L.J.; Hong, Q.; Schultz, L.; Kuo, E.; Lin, S.R.; Lee, M.H.; Lin, Y.S.; Chang, N.S. Zfra is an inhibitor of Bcl-2 expression and cytochrome c release from the mitochondria. Cell. Signal. 2008, 20, 1303–1312.
More
