The HSP90 paralog TRAP1 was discovered more than 20 years ago, however, a detailed understanding of the function of this mitochondrial molecular chaperone remains elusive. The dispensable nature of TRAP1 in vitro and in vivo further complicates an understanding of its role in mitochondrial biology. TRAP1 is more homologous to the bacterial HSP90, HtpG, than to eukaryotic HSP90. Lacking co-chaperones, the unique structural features of TRAP1 likely regulate its temperature-sensitive ATPase activity and shed light on the alternative mechanisms driving the chaperone’s nucleotide-dependent cycle in a defined environment whose physiological temperature approaches 50 °C. TRAP1 has been shown to be an important bioregulator of mitochondrial respiration, mediating the balance between oxidative phosphorylation and glycolysis, while at the same time promoting mitochondrial homeostasis and displaying cytoprotective activity.
1. Molecular Cchaperones, HSP90 and TRAP1
Molecular chaperones form one of the central pillars of the cellular proteostasis network
[1][2][1,2]. Depending upon their function, these molecules fall into three fundamental classes: foldases, holdases, and disaggregases
[1][2][3][4][5][6][7][1,2,3,4,5,6,7]. Under certain circumstances, some molecular chaperones also deliver damaged and impossible to fold client proteins for degradation by proteosomes or autophagy
[8][9][10][8,9,10]. Foldases are ATP-dependent chaperones that actively fold nascent proteins into their native functional conformations and refold unfolded proteins under cellular stress. Heat shock protein 90 (HSP90) is an ATP-dependent foldase that is remarkably conserved from bacteria to humans
[11]. It regulates folding, maturation, and stability of proteins (in HSP90’s case, termed “clients”) that are involved in cell growth, survival, apoptosis, and adaptation to stress
[12][13][14][12,13,14].
In mammalian cells there are four different HSP90 paralogs: HSP90α, HSP90β, GRP94, and TRAP1. HSP90α and HSP90β are primarily cytosolic with a small component in the nucleus. HSP90α is stress induced while HSP90β is constitutively expressed
[15]. GRP94 is localized in the endoplasmic reticulum
[16] and TRAP1 (or HSP75), the paralog on which
rwe
searchers focused in this
entryreview, is primarily localized in mitochondria
[17][18][17,18]. TRAP1 was initially identified in 2000
[17] and was widely presumed to facilitate late-stage folding of clients in the mitochondrial matrix. However, it increasingly became clear that this may not be the case. TRAP1 has since been implicated in metabolic regulation
[19][20][21][22][23][24][25][19,20,21,22,23,24,25], mitochondrial dynamics
[26], mitophagy
[27][28][27,28], protection from oxidative stress
[23][29][30][31][32][23,29,30,31,32], and protection from cell death
[33].
2. Is TRAP1 Ccytoprotective or Pro-Npro-neoplastic?
Although TRAP1 may have regulatory roles in organellar processes, whether it is ultimately cytoprotective in the context of neurodegenerative diseases or pro-neoplastic in the context of many cancers may reflect two sides of the same coin. This molecule has been reported to play a crucial role in inhibiting oxidative stress-induced tissue damage in the ischemic brain
[34], hypoxia-induced injury in cardiomyocytes
[35], myocardial ischemia/reperfusion injury
[36], motor neuron degeneration in oxidative stress-induced amyotrophic lateral sclerosis (ALS)
[37], and acidosis-induced injury in cardiomyocytes
[38]. Likewise, TRAP1 appears to be protective in genetic models of neurodegeneration such as Parkinson’s disease
[27][28][39][27,28,39] where protein quality control in mitochondria plays a critical role
[40]. TRAP1 was also shown to be mitoprotective in models of kidney fibrosis and renal cell carcinoma
[41][42][41,42]. Finally, loss-of-function TRAP1 mutations have been identified in the brain of a patient with Parkinson’s disease
[43], Leigh syndrome
[44], and chronic functional symptomatology including pain, fatigue, and gastrointestinal dysmotility
[45], and in congenital abnormalities associated with the kidney (CAKUT)
[46].
While these studies identify TRAP1 as cytoprotective in mitochondrial-associated neuropathologies, other studies have highlighted a potential pro-neoplastic role of TRAP1 in cancer, where it can also display cytoprotective and other pro-tumorigenic activities. Thus, TRAP1 expression was found to be increased in hepatocellular carcinoma
[47], breast cancer
[48], glioma
[49], small cell lung cancer
[50], and kidney, prostate, ovarian, colorectal, and esophageal cancer, and it is correlated with advanced-stage metastatic tumors with poor prognosis
[51][52][53][54][55][56][57][51,52,53,54,55,56,57]. In colorectal cancer and its animal models, increased TRAP1 expression was found to be localized to pro-neoplastic lesions in the tumor
[58][59][58,59]. While data supporting the importance of TRAP1 are numerous
[24][47][48][49][50][51][52][53][54][55][56][57][59][60][24,47,48,49,50,51,52,53,54,55,56,57,59,60], these findings are challenged by other reports where TRAP1 expression inversely correlates with tumor stage
[19] or is seemingly unimpactful in carcinogenesis models in TRAP1 knockout (KO) mice
[61]. This has led to a general consensus that TRAP1’s role may be more context dependent.
Nevertheless, TRAP1 does appear to play a role in the metabolic adaptation that may sustain neoplastic growth in a nutrient- and oxygen-poor environment. This hypothesis has driven research to mechanistically elucidate a role played by TRAP1 that is common to various cancers. Thus, TRAP1 was reported to play a critical role in the metabolic switch from oxidative phosphorylation (OxPhos) to aerobic glycolysis
[19]. This relationship of TRAP1 to metabolic plasticity sparked an interest in exploring the details of TRAP1 structure, interactome, mode of action, and inhibitors. The data that has emerged since has definitively highlighted TRAP1 as a major player in mitochondrial bioenergetics.
3. TRAP1 Sstructure, ATPase Ccycle, Ddimers, and Ttetramers
The TRAP1 gene is evolutionarily conserved
[62] and is found in both metazoans and protozoans but not in the budding yeast. Unlike HSP90, TRAP1 is not an essential protein, and TRAP1 KO mice or cells derived therefrom are viable
[19][63][19,63]. Likewise, loss of TRAP1 function in a patient with Parkinson’s disease was unimpactful
[43]. Similar to all members of the HSP90 family, TRAP1 has been primarily reported to form and function as a homodimer, with each protomer being comprised of an N-terminal ATPase domain (NTD), a middle domain (MD), and a C-terminal dimerization domain (CTD)
[11][64][65][11,64,65]. The N-terminal domain contains a 59-amino acid mitochondrial-targeting sequence that is cleaved upon import
[66]. Interestingly, TRAP1 more closely resembles bacterial HSP90 (HtpG) than human HSP90
[17][67][17,67]. As with HtpG, but unlike HSP90, TRAP1 lacks both a charged linker domain between the NTD and MD and a C-terminal EEVD motif that serves as a co-chaperone interaction domain in HSP90. TRAP1 also features an extended β-strand in the NTD, called “strap”, that facilitates a cross protomer interaction in
trans in the closed state of TRAP1. Removal of the “strap” domain dramatically upregulates ATPase activity; this extension is considered to be involved in the thermoregulation of the TRAP1 ATPase and to be potentially inhibitory for TRAP1 function under low temperatures
[68].
TRAP1 is a nucleotide-dependent and nucleotide-activated chaperone that exists as a coiled-coil dimer in an autoinhibited state in the absence of ATP
[69]. The presence of ATP activates the TRAP1 homodimer, which cycles between an open “apo” state and a closed state involving a series of ATP-dependent steps that promote large conformational changes within the molecule
[70]. Unlike the rest of the HSP90 family, TRAP1 has a unique ATP-bound catalytically active state that adopts a strained asymmetric conformation
[71]. This unique asymmetry is most pronounced in the highly conserved client binding region and results from the buckling of one of the protomers onto the other
[71]. Interestingly, ATP hydrolysis is sequential between the two protomers, with the dimer undergoing a “flip” in the asymmetry while still remaining in the closed state
[71]. The first ATP hydrolysis step facilitates client folding while the second leads to client unloading and return to an apo state
[68]. The Mg
2+ ion is the primary choice of cofactor for the TRAP1 ATPase, but it can be replaced by Ca
2+ [72]. Surprisingly, Ca
2+-bound TRAP1 displays cooperative ATP hydrolysis and avoids asymmetric flipping of its protomers
[72]. This may indicate that TRAP1 can function both as a foldase and a holdase, depending on its ionic environment.
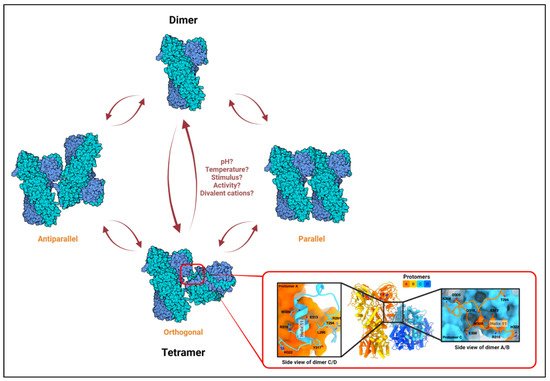
Recently, TRAP1 was reported to form tetramers (dimer of dimers)
[22], and it was proposed that the TRAP1 molecule exists in a dynamic equilibrium between a dimeric and a tetrameric state within mitochondria
[22]. Analytical ultracentrifugation (AUC) with recombinant proteins further confirmed the existence of TRAP1 tetramers, which also seem to be stabilized in vitro by AMPPNP
[73], a non-hydrolyzable structural homolog of ATP. Finally, cryo-EM analyses with purified proteins showed that the TRAP1 tetramer may adopt an orthogonal (butterfly), parallel, or antiparallel conformation (
Figure 1)
[73]. It should be noted that these observations are recent; any functional relevance of TRAP1 tetramers or for the potential transition between configurations remains unknown. Nevertheless, these observations are not entirely surprising when considering that crystallization of bacterial HtpG found the chaperone to exist as a dimer of dimers
[74]. Similarly, HSP90 has also been reported to form such “oligomers”
[75][76][75,76] under certain stimuli including elevated temperatures
[77][78][79][77,78,79] and in the presence of non-ionic detergents or divalent cations
[78][80][78,80].
Figure 1. The TRAP1 tetramer. Based on in vitro studies on HSP90 oligomerization, rapid alterations in temperature, chaperone activity, or local concentration of divalent cations, which are common occurrences in the mitochondria, may influence dimer–tetramer transition. Three distinct conformations have been observed for the TRAP1 tetramer in vitro: orthogonal, parallel, and antiparallel
[22][73][22,73]. The conditions required for the adoption of or transition to a particular configuration are only predicted and remain unclear. A high-resolution MD-MD dimer–dimer interface has only been shown for the orthogonal structure (shown in the inset; adapted from Liu et. al., Biorxiv., 2020
[73]). Left and right sub-insets show interacting residues from protomer C (blue) to A (orange) and from A to C at the dimer–dimer interface, respectively.
Temperature-induced oligomerization of HSP90 is of particular interest in the context of TRAP1. This is because mitochondria operate close to 50 °C under physiological conditions, which is much higher than the 37 °C that is maintained in the adjacent cytosol
[81]. To understand a physiological role of temperature-induced HSP90 oligomers, one study showed that self-oligomerized HSP90 under higher temperatures (>46 °C) readily binds to chemically unfolded dihydrofolate reductase (DHFR), a protein that could spontaneously refold by itself, to maintain it in a “folding-competent” state
[79]. The binding of such a quaternary structure formed by HSP90 may actually provide an ideal environment for protein accommodation prior to folding and is consistent with a holdase function
[82]. This hypothesis, while intriguing, definitely needs further experimental support. In the case of TRAP1, the existence of tetramers in “hot” mitochondria, the alterations in its asymmetry based on the availability of Mg
2+ or Ca
2+ ions, and a lack of significant proteome imbalance in TRAP1 KO cells
[22] are consistent with the ability to adopt a holdase function in the mitochondrial environment. Additional experiments are needed to support or refute this hypothesis.