You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Chundong Yu.
Histone demethylase JMJD2D is a multifunctional epigenetic factor coordinating androgen receptor activation, DNA damage repair, DNA replication, cell cycle regulation, and inflammation modulation. JMJD2D is also a well-established epigenetic facilitator in the progression of multiple malignant tumors, especially in colorectal cancer (CRC) and hepatocellular cancer (HCC).
- JMJD2D
- KDM4D
- H3K9me3
1. Introduction
Genetic materials are tightly condensed in a core of positively charged histones that surround negatively charged DNA wraps. The theory of epigenetics was first proposed by Conrad h. Waddington in 1942, which has been recognized as epigenetic modifications mediating cellular phenotype or gene expression through DNA or histone covalent modifications, chromatin remodeling, non-coding RNA, etc., without altering the sequence of DNA [1,2][1][2]. The core histones constituting nucleosomes contain a Lys- and Arg-rich tail or side chain and are extensively modulated by posttranslational modifications (PTMs) [3]. The acetylation, methylation, and phosphorylation modifications of histone have been widely reported, while it can also be modified by other means such as O-acetyl glycosylation, formylation, and ADP-ribosylation [4,5,6,7,8][4][5][6][7][8].
Cancer progression is closely related to the changes in histone PTMs, and the abnormal control of PTMs-related enzymes, including histone methylase, demethylase, acetylase, and acetyltransferase, is an important predisposing factor for tumors [9]. These epigenetic changes may silence multiple tumor suppressor genes or activate oncogenes, leading to the reprogramming of oncogenes in the genome. Allfrey et al. first reported that histone methylation is related to gene transcriptional regulation [10], and then the modifications on Lys, Arg, Ser, Thr, Tyr, and His residues have been reported [11]. The most widely studied modified site is Lys, which can be monomethyl, dimethyl, or trimethyl (i.e., me1, me2, or me3). There is a relatively large lysine methyltransferase family [12]; however, the same type of methylation modification cannot be catalyzed by all enzymes, which have substrate sequence preference. For example, EZH2 can mediate the monomethylation, dimethylation, and trimethylation of H3K27, while G9a (EHMT2) and MMSET (NSD2) mediate the monomethylation and dimethylation of H3K9 and H3K36, respectively, but not trimethylation [13].
Histone methylation modification was once considered to be an irreversible process, which is overturned by the discovery of LSD1 and KDM demethylase family with Jumonji C (JmjC) domain [14,15,16][14][15][16]. Unlike LSD1, KDMs are oxygenases that target multiple sites including H3K9, H3K27, and H3K36, and utilize 2-ketoglutarate (2-OG) and Fe2+ as cofactors to realize the demethylation of substrates through the mechanism of generating formaldehyde-free radicals [17]. The KDM demethylase JMJD2D (also known as KDM4D) belongs to JMJD2 subfamily that contains five members (JMJD2A-E) and can recognize the dimethyl and trimethylation of H3K9 and H3K36, as well as the trimethylation of H1.4K26 [9,18][9][18]. H3K9 and H1.4K26 trimethylations are related to transcriptional inhibition or heterochromatin formation, while H3K36 methylation is related to the activation of genes; and methylation modification at these sites is not only involved in gene transcription control but also related to DNA replication and repair [19,20][19][20]. Therefore, the dynamic transition of histone methylation regulated by the JMJD2 family may have a far-reaching impact on various cellular physiological activities, especially on tumorigenesis. The research on the function of JMJD2A, JMJD2B, and JMJD2C is relatively comprehensive, while that of JMJD2D is ongoing, but JMJD2E is unformed.
2. JMJD2D Promotes Gene Transcription by Antagonizing H3K9 Methylation
KDM4/JMJD2 histone demethylase family members are very similar in overall protein structure, which contains the featured JmjN and JmjC domain. JMJD2A, JMJD2B, and JMJD2C contain two plant homeodomains (PHD) and two tudor domains, while JMJD2D and JMJD2E are smaller and lack of the C-terminal PHD and tudor domains [9,18,21][9][18][21] (Figure 1A). Intriguingly, Shim et al. demonstrated that the truncation mutants of JMJD2A and JMJD2C that lack the tudor or both the tudor and PHD domains can still demethylate H3K9 and H3K36 [22]. Methylation of some lysine residues in H3 histones can activate or inactivate the transcriptional activity of genes, including H3K4, H3K9, H3K17, H3K27, H3K36, and H3K79 [23], while the dimethylation or trimethylation of H3K9 (H3K9me2/3) and H3K27 (H3K27me2/3) primarily associate with heterochromatin and gene repression [24,25,26,27,28,29][24][25][26][27][28][29]. The full-length JMJD2D, which localizes in the human chromosome 11q21 [18[18][30],30], demethylates the histone residues H3K9me2/3 and H1.4K26me3 to the monomethyl state, although H3K9me2/3 is the preferred substrate [9,31][9][31]. The JmjC domain of JMJD2D is the catalytic core, which facilitates a dioxygenase reaction requiring Fe2+, O2, and 2-oxoglutarate (2-OG) to demethylate histones. The oxygen atom is incorporated into the methyl group during oxidative decarboxylation; subsequently, an unstable intermediate imine product is formed and then converted to formaldehyde and demethylated product (Figure 1B) [32]. As expected, the JmjC-domain-lacking JMJD2 protein is deficient in demethylation activity [22]. The histidine 192 on the JmjC domain of JMJD2D is essential for its histone demethylase function and mutation of this residue severely compromises the catalytic activity of JMJD2D, leading to the loss of its ability to stimulate the mouse mammary tumor virus (MMTV) promoter [33]. The serine 200 on the JmjC domain is also essential for the histone demethylase function of JMJD2D and mutating this residue to methionine (S200M) generates a demethylase-dead mutant [34,35][34][35]. The JmjN domain of JMJD2D may be responsible for the integrity of structure and serves as a dimerization interface [9,18,21,36][9][18][21][36]. Deletion of the JmjN domain also abrogates the histone demethylase activity in the cell, due to the nuclear exclusion of JMJD2D [22].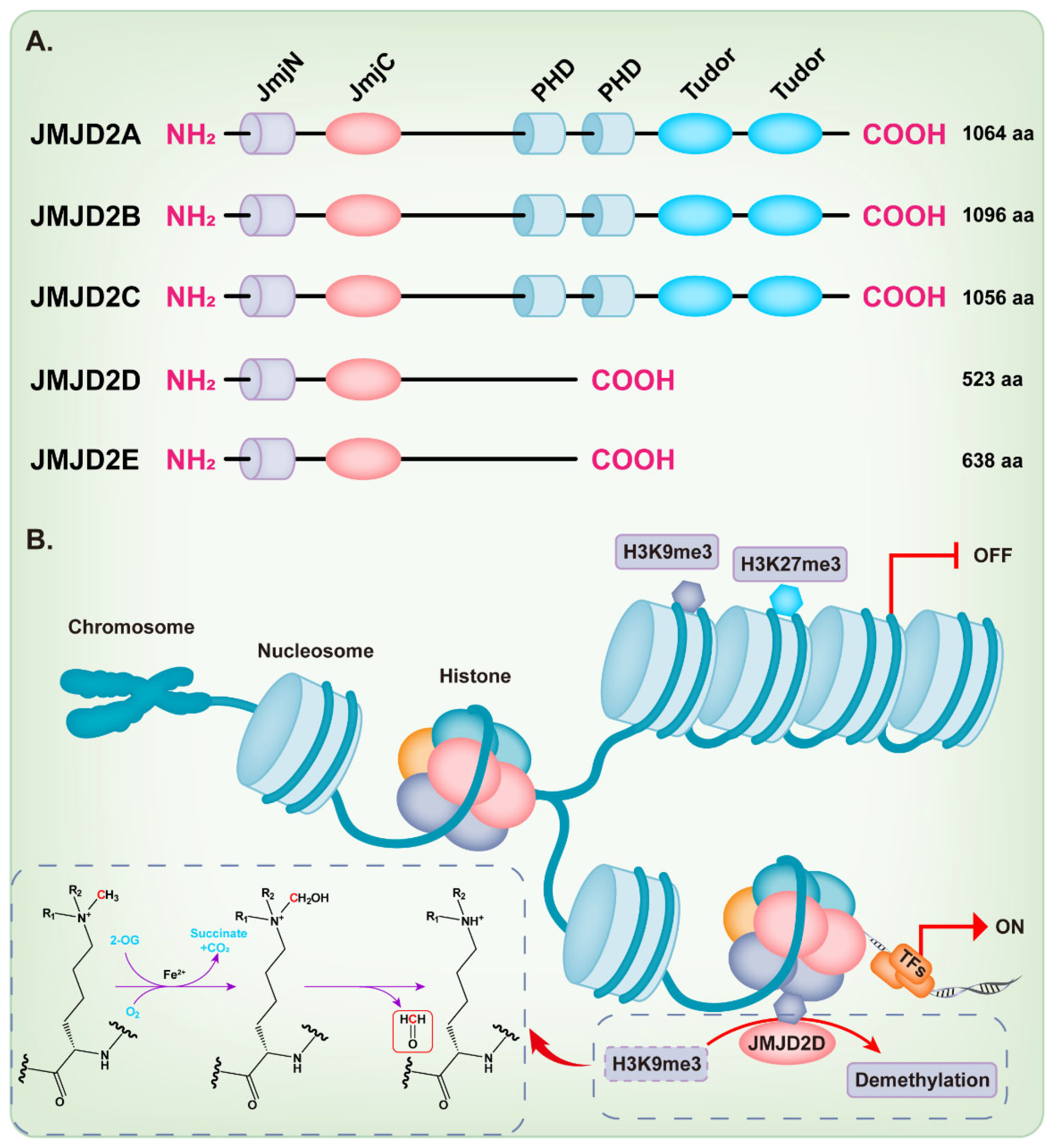
Figure 1. Structure and function of the JMJD2 histone demethylase subfamily. (A) The domain structure diagram of the JMJD2 histone demethylase subfamily. (B) JMJD2D facilitates a dioxygenase reaction requiring Fe2+, O2, and 2-oxoglutarate (2-OG) to demethylate H3K9me3.
3. JMJD2D Has Multiple Biological Functions
As mentioned earlier, JMJD2D promotes gene transcription by antagonizing H3K9 methylation, while it can serve as a coactivator to enhance the transcriptional activities of multiple transcription factors (TFs). JMJD2D is a well-established epigenetic regulator in a variety of biological functions, including androgen receptor (AR) activation, DNA damage repair, DNA replication, cell cycle regulation, inflammation modulation, and tumorigenesis promotion (Figure 2).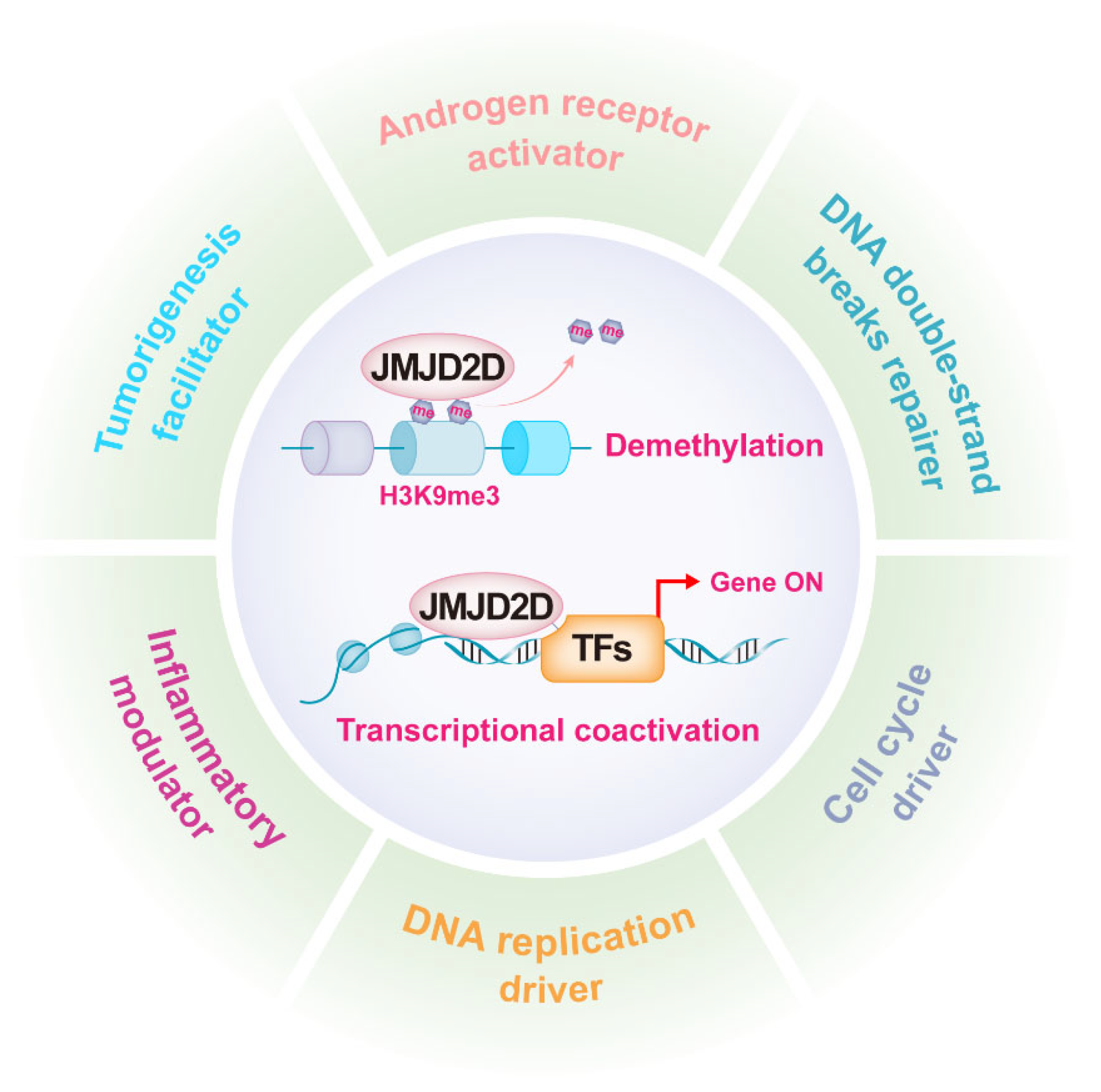
Figure 2. JMJD2D is a multifunctional epigenetic regulator. JMJD2D can promote gene transcription by antagonizing H3K9 methylation and cooperating with multiple transcription factors. JMJD2D has a variety of biological functions, including AR activation, DNA damage repair, DNA replication, cell cycle regulation, inflammation modulation, and tumorigenesis promotion.
3.1. JMJD2D Facilitates the Functions of Androgen Receptor
JMJD2D first attracted attention as an AR activator [33], and the study of its biological function has been ongoing since it was identified. AR is a transcription factor that plays a pivotal role in the development of prostate cancer. Shin and colleagues first reported that JMJD2D is a novel AR coactivator, which can form a complex with ligand-bound AR via its C-terminus to promote the progression of prostate cancer [33]. Androgen is also an important regulator for trophoblast differentiation and placental development, JMJD2D facilitates this process via forming the AR-KDMs complex and promoting the placental androgen signaling to regulate placental VEGFA expression [37]. JMJD2D, which is highly expressed in testis and strongly associated with testis morphology traits [38], demethylates H3K9me3 during spermatogenesis in mice; however, the fertility of JMJD2D-deficient mice is not undulated although H3K9me3 accumulates significantly in round spermatids [39].3.2. JMJD2D Plays a Key Role in DNA Damage Repair, DNA Replication, and Cell Cycle Regulation
DNA damage is one of the hallmarks of cancer that leads to genomic instability, in which the double-strand breaks (DSB) are the most deleterious damage forms and H3K9me3 is one of the epigenetic barriers to DSB repair [40,41][40][41]. Previously, JMJD2B demethylase was identified as a DNA damage response protein, which can be recruited to DNA damage tracks induced by laser micro-irradiation in a demethylase-dependent manner [40]. Khoury-Haddad et al. also reported that JMJD2D demethylase, which can be rapidly recruited to DNA damage regions, is required for DSB repair, while the demethylase activity of JMJD2D is dispensable for the accumulation at DNA damage regions, but its C-terminal region is essential [34]. Intriguingly, Khoury-Haddad et al. demonstrated in another study that JMJD2D–RNA interaction is required for the recruitment of JMJD2D to DNA damage sites, and the efficient demethylation of H3K9me3 by JMJD2D is essential [42]. H3K9me3 is associated with transcriptional silencing in heterochromatin, while it can also inactivate the enhancers of the cell-type-specific gene to participate in the precise regulation of gene expression, and the demethylation of H3K9me3 by JMJD2D demethylase is essential in this process [43,44][43][44]. Furthermore, sufficient evidence showed that JMJD2D is involved in DNA replication and cell cycle regulation [45,46,47][45][46][47]. H3K9me3 is associated with the silencing of cell cycle genes that are essential for cardiac myocyte (CM) cell cycle exit; while overexpression of JMJD2D specifically attenuates H3K9me3 levels and increases the expression of cell-cycle-associated proteins in CM, resulting in CM hyperplasia [47]. Again, Wu et al. reported that JMJD2D combines with DNA replication-related proteins via its JmjC domain in a demethylation activity independent manner during G1 and S phases of the cell cycle, which promotes the initiation and elongation of DNA replication [45]. Although the binding of JMJD2D does not depend on its demethylase activity, reducing the level of H3K9me3 can rescue the DNA replication defect in JMJD2D-deficient cells, indicating that the demethylase activity of JMJD2D may be essential [45].3.3. JMJD2D Modulates the Inflammatory Response
In addition to the above-mentioned JMJD2D-modulated biological activities, other studies have shown that JMJD2D may be involved in immune and inflammatory responses, and it also plays a critical role in the neurogenesis of dynamic hippocampal dentate gyrus [48,49][48][49]. Jin and colleagues reported that JMJD2D is involved in the regulation of proinflammatory cytokines interleukin 12 and 23 (IL-12 and IL-23), which are regulators of immune response and inflammation [48]. TRAF-binding protein domain (Trabid, also known as Zranb1), a deubiquitinase that preferentially hydrolyzes lysine 29 (K29)- and K33-linked ubiquitin chain [50[50][51],51], is a crucial regulator of TLR-stimulated IL-12 and IL-23 expression [48]. LPS stimulation upregulates the expression of JMJD2D and promotes its recruitment to the IL12a, IL12b, and IL23a promoters [48,52][48][52]. Trabid effectively cooperates with JMJD2D to promote the expression of these inflammatory factors by preferentially reducing the K29 ubiquitination of JMJD2D and stabilizing JMJD2D [48]. The autophagic degradation of JMJD2D in modulating inflammatory response has also been reported. Tripartite motif 14 (TRIM14), which belongs to one of the most dominant E3 ligase families, is a critical regulator of type I IFN signaling [53] and an important oncogene [54,55][54][55]. TRIM14 decreases the H3K9me2/3 level to facilitate the transcription of IL-12 and IL-23 by reducing the K63-linked ubiquitination of JMJD2D and subsequently prevents its selective autophagic degradation [56].References
- Waddington, C.H. Canalization of development and genetic assimilation of acquired characters. Nature 1959, 183, 1654–1655.
- Okugawa, Y.; Grady, W.M.; Goel, A. Epigenetic Alterations in Colorectal Cancer: Emerging Biomarkers. Gastroenterology 2015, 149, 1204–1225.e12.
- Audia, J.E.; Campbell, R.M. Histone Modifications and Cancer. Cold Spring Harb. Perspect. Biol. 2016, 8, a019521.
- Chen, Y.; Sprung, R.; Tang, Y.; Ball, H.; Sangras, B.; Kim, S.C.; Falck, J.R.; Peng, J.; Gu, W.; Zhao, Y. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell. Proteom. 2007, 6, 812–819.
- Martin, C.; Zhang, Y. Mechanisms of epigenetic inheritance. Curr. Opin. Cell Biol. 2007, 19, 266–272.
- Ruthenburg, A.J.; Li, H.; Patel, D.J.; Allis, C.D. Multivalent engagement of chromatin modifications by linked binding modules. Nat. Rev. Mol. Cell Biol. 2007, 8, 983–994.
- Tan, M.; Luo, H.; Lee, S.; Jin, F.; Yang, J.S.; Montellier, E.; Buchou, T.; Cheng, Z.; Rousseaux, S.; Rajagopal, N.; et al. Identification of 67 histone marks and histone lysine crotonylation as a new type of histone modification. Cell 2011, 146, 1016–1028.
- Herranz, N.; Dave, N.; Millanes-Romero, A.; Morey, L.; Díaz, V.M.; Lórenz-Fonfría, V.; Gutierrez-Gallego, R.; Jerónimo, C.; Di Croce, L.; García de Herreros, A.; et al. Lysyl oxidase-like 2 deaminates lysine 4 in histone H3. Mol. Cell 2012, 46, 369–376.
- Berry, W.L.; Janknecht, R. KDM4/JMJD2 histone demethylases: Epigenetic regulators in cancer cells. Cancer Res. 2013, 73, 2936–2942.
- Allfrey, V.G.; Faulkner, R.; Mirsky, A.E. Acetylation and methylation of histones and their possible role in the regulation of RNA synthesis. Proc. Natl. Acad. Sci. USA 1964, 51, 786–794.
- Rothbart, S.B.; Strahl, B.D. Interpreting the language of histone and DNA modifications. Biochim. Biophys. Acta 2014, 1839, 627–643.
- Arrowsmith, C.H.; Bountra, C.; Fish, P.V.; Lee, K.; Schapira, M. Epigenetic protein families: A new frontier for drug discovery. Nat. Rev. Drug Discov. 2012, 11, 384–400.
- Kuo, A.J.; Cheung, P.; Chen, K.; Zee, B.M.; Kioi, M.; Lauring, J.; Xi, Y.; Park, B.H.; Shi, X.; Garcia, B.A.; et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell 2011, 44, 609–620.
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953.
- McAllister, T.E.; England, K.S.; Hopkinson, R.J.; Brennan, P.E.; Kawamura, A.; Schofield, C.J. Recent Progress in Histone Demethylase Inhibitors. J. Med. Chem. 2016, 59, 1308–1329.
- Jambhekar, A.; Anastas, J.N.; Shi, Y. Histone Lysine Demethylase Inhibitors. Cold Spring Harb. Perspect. Med. 2017, 7, a026484.
- Tsukada, Y.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439, 811–816.
- Katoh, M.; Katoh, M. Identification and characterization of JMJD2 family genes in silico. Int. J. Oncol. 2004, 24, 1623–1628.
- Chi, P.; Allis, C.D.; Wang, G.G. Covalent histone modifications--miswritten, misinterpreted and mis-erased in human cancers. Nat. Rev. Cancer 2010, 10, 457–469.
- Dawson, M.A.; Kouzarides, T. Cancer epigenetics: From mechanism to therapy. Cell 2012, 150, 12–27.
- Whetstine, J.R.; Nottke, A.; Lan, F.; Huarte, M.; Smolikov, S.; Chen, Z.; Spooner, E.; Li, E.; Zhang, G.; Colaiacovo, M.; et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 2006, 125, 467–481.
- Shin, S.; Janknecht, R. Diversity within the JMJD2 histone demethylase family. Biochem. Biophys. Res. Commun. 2007, 353, 973–977.
- Lawrence, M.; Daujat, S.; Schneider, R. Lateral Thinking: How Histone Modifications Regulate Gene Expression. Trends Genet. 2016, 32, 42–56.
- Pedersen, M.T.; Kooistra, S.M.; Radzisheuskaya, A.; Laugesen, A.; Johansen, J.V.; Hayward, D.G.; Nilsson, J.; Agger, K.; Helin, K. Continual removal of H3K9 promoter methylation by Jmjd2 demethylases is vital for ESC self-renewal and early development. EMBO J. 2016, 35, 1550–1564.
- Kundaje, A.; Meuleman, W.; Ernst, J.; Bilenky, M.; Yen, A.; Heravi-Moussavi, A.; Kheradpour, P.; Zhang, Z.; Wang, J.; Ziller, M.J.; et al. Integrative analysis of 111 reference human epigenomes. Nature 2015, 518, 317–330.
- Mozzetta, C.; Boyarchuk, E.; Pontis, J.; Ait-Si-Ali, S. Sound of silence: The properties and functions of repressive Lys methyltransferases. Nat. Rev. Mol. Cell Biol. 2015, 16, 499–513.
- Mikkelsen, T.S.; Ku, M.; Jaffe, D.B.; Issac, B.; Lieberman, E.; Giannoukos, G.; Alvarez, P.; Brockman, W.; Kim, T.K.; Koche, R.P.; et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 2007, 448, 553–560.
- Wang, Z.; Zang, C.; Rosenfeld, J.A.; Schones, D.E.; Barski, A.; Cuddapah, S.; Cui, K.; Roh, T.Y.; Peng, W.; Zhang, M.Q.; et al. Combinatorial patterns of histone acetylations and methylations in the human genome. Nat. Genet. 2008, 40, 897–903.
- Vakoc, C.R.; Sachdeva, M.M.; Wang, H.; Blobel, G.A. Profile of histone lysine methylation across transcribed mammalian chromatin. Mol. Cell. Biol. 2006, 26, 9185–9195.
- Taylor, T.D.; Noguchi, H.; Totoki, Y.; Toyoda, A.; Kuroki, Y.; Dewar, K.; Lloyd, C.; Itoh, T.; Takeda, T.; Kim, D.W.; et al. Human chromosome 11 DNA sequence and analysis including novel gene identification. Nature 2006, 440, 497–500.
- Weiss, T.; Hergeth, S.; Zeissler, U.; Izzo, A.; Tropberger, P.; Zee, B.M.; Dundr, M.; Garcia, B.A.; Daujat, S.; Schneider, R. Histone H1 variant-specific lysine methylation by G9a/KMT1C and Glp1/KMT1D. Epigenet. Chromatin 2010, 3, 7.
- Krishnan, S.; Collazo, E.; Ortiz-Tello, P.A.; Trievel, R.C. Purification and assay protocols for obtaining highly active Jumonji C demethylases. Anal. Biochem. 2012, 420, 48–53.
- Shin, S.; Janknecht, R. Activation of androgen receptor by histone demethylases JMJD2A and JMJD2D. Biochem. Biophys. Res. Commun. 2007, 359, 742–746.
- Khoury-Haddad, H.; Guttmann-Raviv, N.; Ipenberg, I.; Huggins, D.; Jeyasekharan, A.D.; Ayoub, N. PARP1-dependent recruitment of KDM4D histone demethylase to DNA damage sites promotes double-strand break repair. Proc. Natl. Acad. Sci. USA 2014, 111, E728–E737.
- Peng, K.; Kou, L.; Yu, L.; Bai, C.; Li, M.; Mo, P.; Li, W.; Yu, C. Histone Demethylase JMJD2D Interacts With β-Catenin to Induce Transcription and Activate Colorectal Cancer Cell Proliferation and Tumor Growth in Mice. Gastroenterology 2019, 156, 1112–1126.
- Levin, M.; Stark, M.; Assaraf, Y.G. The JmjN domain as a dimerization interface and a targeted inhibitor of KDM4 demethylase activity. Oncotarget 2018, 9, 16861–16882.
- Cleys, E.R.; Halleran, J.L.; Enriquez, V.A.; da Silveira, J.C.; West, R.C.; Winger, Q.A.; Anthony, R.V.; Bruemmer, J.E.; Clay, C.M.; Bouma, G.J. Androgen receptor and histone lysine demethylases in ovine placenta. PLoS ONE 2015, 10, e0117472.
- Zhou, T.; Wei, H.; Li, D.; Yang, W.; Cui, Y.; Gao, J.; Yu, T.; Lv, X.; Pan, C. A novel missense mutation within the domain of lysine demethylase 4D (KDM4D) gene is strongly associated with testis morphology traits in pigs. Anim. Biotechnol. 2020, 31, 52–58.
- Iwamori, N.; Zhao, M.; Meistrich, M.L.; Matzuk, M.M. The testis-enriched histone demethylase, KDM4D, regulates methylation of histone H3 lysine 9 during spermatogenesis in the mouse but is dispensable for fertility. Biol. Reprod. 2011, 84, 1225–1234.
- Young, L.C.; McDonald, D.W.; Hendzel, M.J. Kdm4b histone demethylase is a DNA damage response protein and confers a survival advantage following γ-irradiation. J. Biol. Chem. 2013, 288, 21376–21388.
- Hanahan, D. Hallmarks of Cancer: New Dimensions. Cancer Discov. 2022, 12, 31–46.
- Khoury-Haddad, H.; Nadar-Ponniah, P.T.; Awwad, S.; Ayoub, N. The emerging role of lysine demethylases in DNA damage response: Dissecting the recruitment mode of KDM4D/JMJD2D to DNA damage sites. Cell Cycle 2015, 14, 950–958.
- Zhu, Y.; van Essen, D.; Saccani, S. Cell-type-specific control of enhancer activity by H3K9 trimethylation. Mol. Cell 2012, 46, 408–423.
- Smith, S.M.; Kimyon, R.S.; Watters, J.J. Cell-type-specific Jumonji histone demethylase gene expression in the healthy rat CNS: Detection by a novel flow cytometry method. ASN Neuro 2014, 6, 193–207.
- Wu, R.; Wang, Z.; Zhang, H.; Gan, H.; Zhang, Z. H3K9me3 demethylase Kdm4d facilitates the formation of pre-initiative complex and regulates DNA replication. Nucleic Acids Res. 2017, 45, 169–180.
- Martins, N.; Cisneros-Soberanis, F.; Pesenti, E.; Kochanova, N.; Shang, W.; Hori, T.; Nagase, T.; Kimura, H.; Larionov, V.; Masumoto, H.; et al. H3K9me3 maintenance on a human artificial chromosome is required for segregation but not centromere epigenetic memory. J. Cell Sci. 2020, 133, jcs242610.
- El-Nachef, D.; Oyama, K.; Wu, Y.Y.; Freeman, M.; Zhang, Y.; MacLellan, W.R. Repressive histone methylation regulates cardiac myocyte cell cycle exit. J. Mol. Cell. Cardiol. 2018, 121, 1–12.
- Jin, J.; Xie, X.; Xiao, Y.; Hu, H.; Zou, Q.; Cheng, X.; Sun, S.C. Epigenetic regulation of the expression of Il12 and Il23 and autoimmune inflammation by the deubiquitinase Trabid. Nat. Immunol. 2016, 17, 259–268.
- Maitra, S.; Khandelwal, N.; Kootar, S.; Sant, P.; Pathak, S.S.; Reddy, S.; Anapoorna, P.K.; Murty, U.S.; Chakravarty, S.; Kumar, A. Histone Lysine Demethylase JMJD2D/KDM4D and Family Members Mediate Effects of Chronic Social Defeat Stress on Mouse Hippocampal Neurogenesis and Mood Disorders. Brain Sci. 2020, 10, 833.
- Virdee, S.; Ye, Y.; Nguyen, D.P.; Komander, D.; Chin, J.W. Engineered diubiquitin synthesis reveals Lys29-isopeptide specificity of an OTU deubiquitinase. Nat. Chem. Biol. 2010, 6, 750–757.
- Licchesi, J.D.; Mieszczanek, J.; Mevissen, T.E.; Rutherford, T.J.; Akutsu, M.; Virdee, S.; El Oualid, F.; Chin, J.W.; Ovaa, H.; Bienz, M.; et al. An ankyrin-repeat ubiquitin-binding domain determines TRABID’s specificity for atypical ubiquitin chains. Nat. Struct. Mol. Biol. 2011, 19, 62–71.
- Boonmee, A.; Benjaskulluecha, S.; Kueanjinda, P.; Wongprom, B.; Pattarakankul, T.; Palaga, T. The chemotherapeutic drug carboplatin affects macrophage responses to LPS and LPS tolerance via epigenetic modifications. Sci. Rep. 2021, 11, 21574.
- Chen, M.; Meng, Q.; Qin, Y.; Liang, P.; Tan, P.; He, L.; Zhou, Y.; Chen, Y.; Huang, J.; Wang, R.F.; et al. TRIM14 Inhibits cGAS Degradation Mediated by Selective Autophagy Receptor p62 to Promote Innate Immune Responses. Mol. Cell 2016, 64, 105–119.
- Sun, W.; Wang, Y.; Li, D.; Wu, Y.; Ji, Q.; Sun, T. Tripartite motif containing 14: An oncogene in papillary thyroid carcinoma. Biochem. Biophys. Res. Commun. 2020, 521, 360–367.
- Chen, J.; Huang, L.; Quan, J.; Xiang, D. TRIM14 regulates melanoma malignancy via PTEN/PI3K/AKT and STAT3 pathways. Aging 2021, 13, 13225–13238.
- Liu, D.; Zhao, Z.; She, Y.; Zhang, L.; Chen, X.; Ma, L.; Cui, J. TRIM14 inhibits OPTN-mediated autophagic degradation of KDM4D to epigenetically regulate inflammation. Proc. Natl. Acad. Sci. USA 2022, 119, e2113454119.
More
