Grapevine (Vitis vinifera) is one of the main fruit crops worldwide, with near of 7.3 million hectares planted in 2020, but along with its economic relevance, it has been associated with diverse pathogens that affect grapevine yield, fruit, and wine quality, of which powdery mildew is the most important disease prior to harvest. Its causal agent is the biotrophic fungus Erysiphe necator, which generates a decrease in cluster weight, delays fruit ripening, and reduces photosynthetic and transpiration rates. In addition, powdery mildew induces metabolic reprogramming in its host, affecting primary metabolism. Most commercial grapevine cultivars are highly susceptible to powdery mildew; consequently, large quantities of fungicide are applied during the productive season. These pesticide applications have been associated with high exposure to it, and pesticides are associated with health problems, negative environmental impacts, and high costs for farmers. In parallel, consumers are demanding more sustainable practices during food production. Therefore, new grapevine cultivars with genetic resistance to powdery mildew are needed for sustainable viticulture, while maintaining yield, fruit, and wine quality. Two main gene families confer resistance to powdery mildew in the Vitaceae, Run (Resistance to Uncinula necator) and Ren (Resistance to Erysiphe necator), and the resistance they confer is associated with the presence of each locus since there are still no genes that alone can produce a powerful genetic resistance. Because the resistance mediated by the plant immune response is highly complex and considers the evolution and adaptation of the pathogen in parallel to that of the plant.
- Erysiphe necator
- grapevine
- resistance genes
- Run
- Ren
- powdery mildew
- Vitis
- Resistance to pathogens
1. Introduction
2. Host Response
The plant’s immune system is summarized by the zig-zag model, which distributes the plant’s response to the presence of pathogens into three main stages. The initial stage is related to the recognition of Pathogen-Associated Molecular Patterns (PAMPs) or Microbe-Associated Molecular Patterns (MAMPs) by Pattern Recognition Receptors (PRRs), resulting in PAMP-triggered immunity (PTI). This triggers nonspecific physiological and molecular responses, such as the accumulation of reactive oxygen species (ROSs) and phytoalexins, and/or stomata closure through the phosphorylation of a MAP kinase pathway (MAPKKK–MAPKK–MAPK), which activates transcription factors, such as WRKY22, thus inducing related genetic responses. In the second stage, and in response to plant defense, pathogens initiate effector-triggered susceptibility (ETS) [29] whereby through effectors (Secreted Effector Proteins), such as coat proteins [30] or other specific proteins, the defensive response pathway of plants is stopped. For instance, some effectors, such as AvrPto and AvrPtoB, have been shown to block the phosphorylation of MAPKs in the case of Pseudomonas syringae [31], while EqCSEP01276 produced by powdery mildew inhibits the biosynthesis of abscisic acid (ABA) [32]. In this ETS phase, pathogens may overcome the immune response of plants and infect the host’s cells. Cells of certain plant species possess resistance proteins (R) that directly or indirectly recognize the presence of pathogenic effectors and trigger an immune response, called effector-triggered immunity (ETI). This final phase generates an immune response of greater intensity than PTI. This switch between ETS–ETI is maintained until the hypersensitive cell death response is triggered or the pathogen overwhelms the cell [33]. Most R genes encode nucleotide-binding site (NBS) leucine-rich repeat (LRR) domain proteins (NBS–LRR proteins) [29]. This is the case of the R genes transcribed in the Vitaceae plant family in response to E. necator infection. In Vitaceae, the R genes are clustered in tandem repeats of genomic regions. These have been genetically mapped, uncovering nine loci that encode R gene sequences conferring resistance to E. necator, such as Run1, Run2, Ren1, Ren2, Ren3, Ren4, Ren5, Ren6, and Ren7 [34], which have been used to obtain plants resistant to this infection by pseudo-backcrossing [35]. On the other hand, more recent “New Breeding Technologies” (NBTs) have been employed for genetic improvements in Vitis plants through the elimination of the endogenous genetic material using the thermal shock FRP/FLP system [36,37][36][37] or the generation of DNA-free modifications using ribonucleoproteins [38]. This, together with new rapidly developing Vitis models, such as Microvine or Picovine, have helped to accelerate the discovery of new target genes to decipher the resistance of Vitis to powdery mildew, such as the PATHOGENESIS-RELATED 4b (VvPR4b) gene, whose loss of function decreases Vitis resistance to downy mildew [39]. As expected, the overexpression of VvPR4b is related to enhanced resistance to E. necator [40], while the DIMERIZATION PARTNER-E2F-LIKE 1 (VviDEL1) double-cut transgenic Vitis has 90% fewer symptoms of powdery mildew infection than the control plants [40]. Hormones play a key role in plant defense responses, particularly jasmonic acid (JA) and ethylene (Et) for necrotrophic pathogens and salicylic acid (SA) for hemibiotrophic and biotrophic pathogens, such as powdery mildew [3,8][3][8]. In Arabidopsis thaliana, SA is synthesized in response to a pathogen attack, mainly from chorismic acid by the activity of the enzymes isochorismate synthase (ICS) and isochorismate pyruvate lyase (IPL) [41]. A mobile derivative of SA is methyl salicylate (MeSa), which can be transported through the phloem to distal parts of plants, generating a Systemic Acquired Response (SAR). This activates various physiological immune responses, such as programmed cell death (PCD) and accumulation of ROS, such as hydrogen peroxide and nitric oxide [42]. Thus, to achieve an effective resistance response in grapevines upon infection by E. necator, it is necessary to enhance SAR [3]. Although the most well-described hormonal response pathway against the attack of powdery mildew is that of SA, it has also been shown that Et and JA contribute to the response against E. necator in grapevines [43,44][43][44]. Furthermore, recent data show that when V. vinifera cv. ‘Cabernet Sauvignon’ plants are treated with exogenous Et, a defense response against E. necator is triggered [44]. Such a response mechanism is associated with the induction of a series of defense proteins, such as acidic class IV chitinase (CHIT4c), protease inhibitor (PIN), polygalacturonase-inhibiting protein (PGIP), and ß-1,3-glucanase (GLU). Although there is no direct evidence linking the induction of these defense proteins with the phenylpropanoid pathway, a correlation has been seen in the increased biosynthesis of phytoalexins and the upregulation of phenylalanine ammonia-lyase (PAL) and stilbene synthase (STS) genes. These increases are positively correlated with the increased accumulation of stilbenes with known antimicrobial activity, which emphasizes the participation of these enzymes in the host response against biotrophic fungi [45]. In support of the above, the transcriptomic analysis of the response to E. necator infection of two Vitis species, one susceptible (V. pseudoreticulata) and the other resistant (V. quinquangularis), showed the induction of genes and metabolites associated with the defense response [46]. Specifically, the repression of the flavonoid pathway genes was reported in the susceptible cultivar V. pseudoreticulata, alongside differential responses of genes and processes related to hormones, such as SA and JA [47]. A high accumulation of arachidic acid has been reported in berries infected by E. necator, meaning that it is now considered a quantitative biomarker for infection by this fungus [8,48][8][48]. Interestingly, Jiao et al. [46] described the suppression of genes related to the biosynthesis and elongation of fatty acids in the resistant cultivar, suggesting the participation of these types of lipids in the interaction of E. necator with the host in a developing infection. Additionally, genes involved in the biosynthesis and signaling of phytohormones, such as JA and cytokinins (CK), were identified, as were ones that code for protein kinases and proteins with NBS–LRR repeats [46][46] (Figure 1).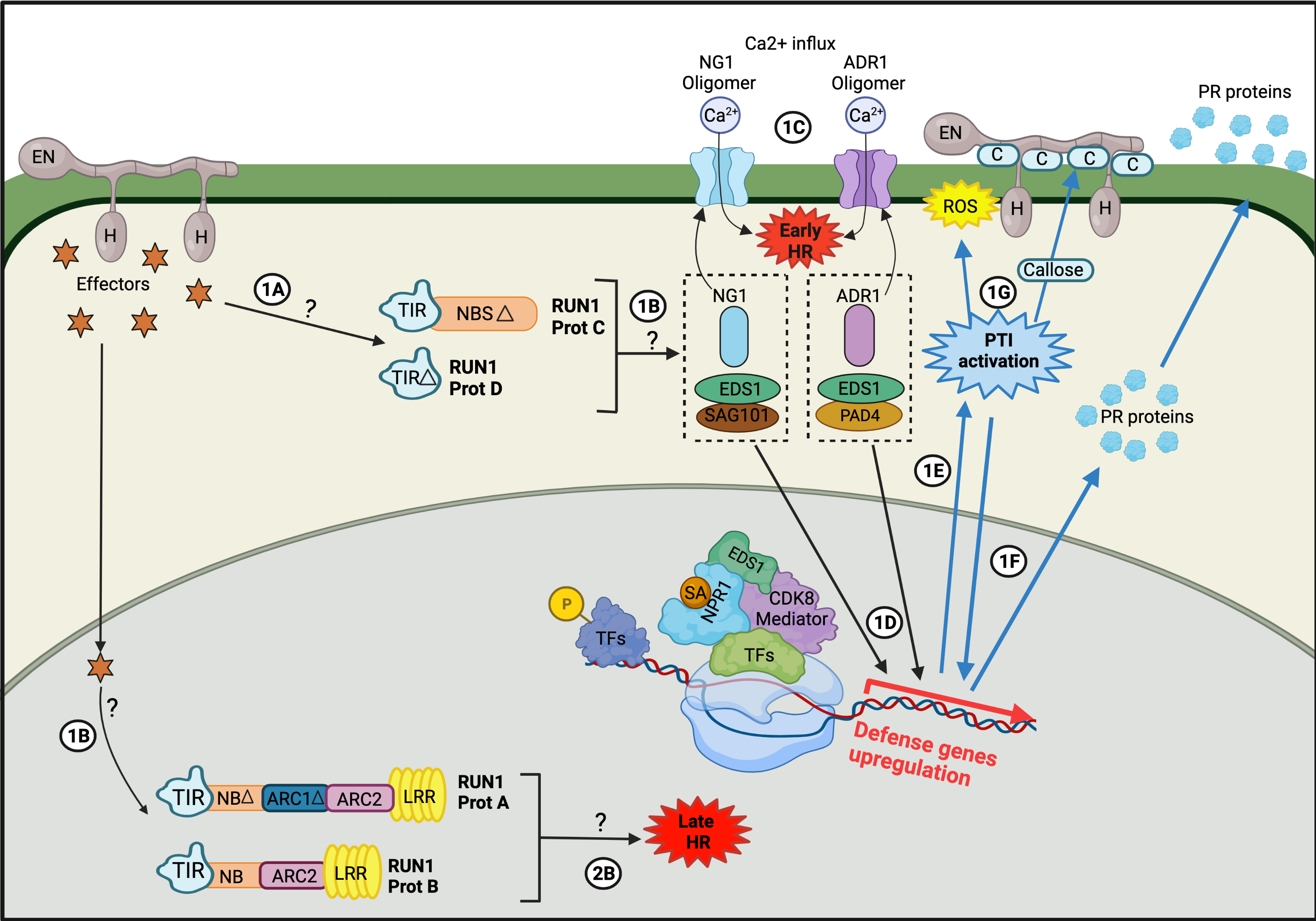 Figure 1. Illustrative figure adapted to summarize the host (V. vinifera) response to fungal (E. necator), showing a theoretical pathway for RUN1: Run1 proteins recognize pathogen effectors, which activate PTI. It is proposed that Run1 proteins modulate two response pathways, one generated by truncated proteins (Prot C and Prot D, pathway1) located in the cytoplasm, and another produced by full-length RUN1 proteins in the cell nucleus (Prot A and Prot B, pathway 2). Pathway 1: 1A) Truncated RUN1 proteins detect fungal effectors in the cytoplasm. 1B) Activation of signaling nodes of PTI: EDS1-PAD4-ADR1 and EDS-SAG10-NGR1. 1C) Activation of Ca2+ influx, which culminates with an early PCD. 1D) EDS1 acts as a transcriptional factor that recruits CDK8 and RNA polymerase II to start the expression of defence genes. 1E) The activation of ETI triggers PTI (blue arrows). 1F) PTI generates the transcription of Plant-pathogenesis Related Proteins (PR proteins). 1G) PTI increases the ROS production and produces callose deposits (C) in the cell areas where the fungus penetrated. Pathway Pathway 22: 1B) E. necator effectors move to the cell nucleus, recognized by the full-length And B RUN1 proteins. 2B) Prot A and prot B triggers a late HR. This illustrative figure was made specifically for this publication by Viviana Sosa-Suñiga, it is adapted to summarize the response pathways to fungi in plants. This representation is general and can only be used as a guide.
Figure 1. Illustrative figure adapted to summarize the host (V. vinifera) response to fungal (E. necator), showing a theoretical pathway for RUN1: Run1 proteins recognize pathogen effectors, which activate PTI. It is proposed that Run1 proteins modulate two response pathways, one generated by truncated proteins (Prot C and Prot D, pathway1) located in the cytoplasm, and another produced by full-length RUN1 proteins in the cell nucleus (Prot A and Prot B, pathway 2). Pathway 1: 1A) Truncated RUN1 proteins detect fungal effectors in the cytoplasm. 1B) Activation of signaling nodes of PTI: EDS1-PAD4-ADR1 and EDS-SAG10-NGR1. 1C) Activation of Ca2+ influx, which culminates with an early PCD. 1D) EDS1 acts as a transcriptional factor that recruits CDK8 and RNA polymerase II to start the expression of defence genes. 1E) The activation of ETI triggers PTI (blue arrows). 1F) PTI generates the transcription of Plant-pathogenesis Related Proteins (PR proteins). 1G) PTI increases the ROS production and produces callose deposits (C) in the cell areas where the fungus penetrated. Pathway Pathway 22: 1B) E. necator effectors move to the cell nucleus, recognized by the full-length And B RUN1 proteins. 2B) Prot A and prot B triggers a late HR. This illustrative figure was made specifically for this publication by Viviana Sosa-Suñiga, it is adapted to summarize the response pathways to fungi in plants. This representation is general and can only be used as a guide.
3. Mapping Resistance Genes for Powdery Mildew Resistance Using Interspecific Crosses
The use of F1 families derived from the cross of two parents with contrasting phenotypes is the most used strategy for genetic mapping in grapevines [49]. Based on the pseudo-testcross strategy [50], it is suitable for highly heterozygous plants with long juvenile periods, such as grapevines. Although V. vinifera is the most widely cultivated Vitis species, the levels of powdery mildew resistance in this species are lower than that of other Vitis or Muscadinia species from North America or Asia. These contrasting phenotypes have been exploited for genetic mapping. To date, 15 loci responsible for grapevine powdery mildew resistance have been reported, leveraging information from 24 F1 interspecific families or descendants [51]. Strong disease-resistant loci have been mapped to chromosomes 12, 18, and 9, named Run1 [52[52][53],53], Ren4 [54[54][55][56],55,56], and Ren6 [57], respectively. These loci originate from M. rotundifolia, V. romanetii, and V. piasezkii and provide strong quantitative disease resistance [58]. Other moderate to minor disease-resistant sources have been found on chromosomes 2, 9, 13, 14, 15, and 18 [51]. In some of these loci, the study of the infection process demonstrated a postpenetration resistance mechanism, with delayed hyphal growth, as in the case of Ren1 [59], Ren5 [60], Ren7 [57], and Ren11 [61,62][61][62]. Some of these moderate to minor resistance loci come from V. vinifera, V. rotundifolia [60], V. piasezkii [57], and complex hybrids involving V. cinerea, V. rupestris, or ‘Seibel’ selections [63,64,65,66,67][63][64][65][66][67].4. Run and Ren Resistance Loci
Several loci associated with powdery mildew resistance have been identified in different species of the Vitaceae family. These loci have been named Ren1 [59], Ren1.2 [68], Ren2 [63[63][69],69], Ren3 [65,70][65][70], Ren4 [54], Ren5 [60], Ren6 [57], Ren7 [57], Ren8 [66], Ren9 [65], Ren10 [67], Ren11 [61], Run1 [52[52][53],53], Run1.2a and b [71], Run2.1 [55[55][71],71], and Run2.2 [55,71][55][71] (Figure 12). In the case of most Run and Ren loci, it is not clear which genes are responsible for powdery mildew resistance and their mechanism of action [34]. The only exception to this is the resistance gene MrRUN1 (MrRGA10), whose sequence was described by Feechan et al. (2013) [53]. The MrRUN1 gene encodes an NBS–LRR resistance protein containing a Toll/interleukin-1 receptor-like (TIR) domain, which recognizes pathogen effectors, thus triggering the hypersensitive response (HR), which is characterized by an increase in ROS production leading eventually to programmed cell death (PCD) in infected cells [69]. The same defense response has been seen in grapevine plants that carry the Run1, Run1.2a, Run1.2b, Run2, Ren1, Ren2, Ren3, Ren4, Ren5, Ren6, Run7, or Ren9 loci (Table 1). These facts suggest that the immune response generated by these loci is mediated by resistance proteins that recognize E. necator effectors and activate ETI [34]. This hypothesis is supported by the presence in other species of resistance genes to powdery mildew that encode for NBS–-LRR proteins [72,73,74,75,76,77,78,79][72][73][74][75][76][77][78][79].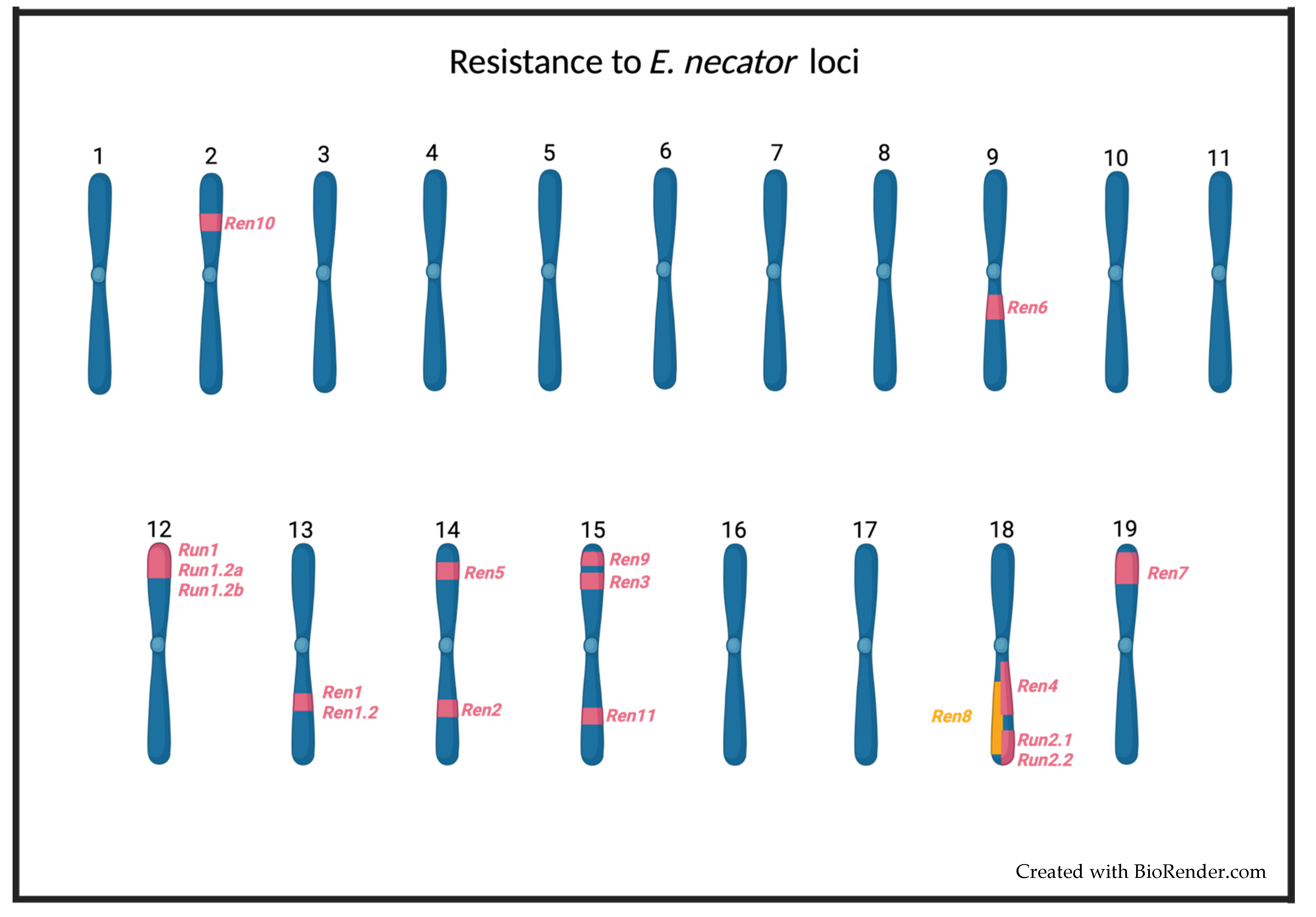
|
Locus |
Donor |
Host Response |
||
|---|---|---|---|---|
|
Effect Type |
Loci | Resistance Level |
Reference |
Reference |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
PCD |
Callose |
ROS |
|||||||||||
|
Run1 |
M. rotundifolia G52 1 |
||||||||||||
|
Additive |
Run1Run1.2a/b |
Yes |
Yes |
Yes |
[69 |
Variable * |
] | ||||||
|
Run1.2a |
M. rotundifolia1 |
Yes |
|||||||||||
|
Run1Ren1 |
[35] | n.i. |
n.i. |
Variable * |
[71] |
||||||||
|
Run1.2b |
M. rotundifolia1 |
Yes |
n.i. |
n.i. |
Variable * |
[71] |
|||||||
|
Run2.1 |
M. rotundifolia ‘Magnolia’ 1 |
Yes |
n.i. |
n.i. |
Partial |
[55] |
|||||||
|
Run2.2 |
M. rotundifolia ‘Trayshed’ 1 |
Yes |
n.i. |
n.i. |
Partial * |
[55] |
|||||||
|
Run1Ren2 * |
[69] |
||||||||||||
|
Nonadditive |
Run1.2a/bRun2.2 |
[69] |
Ren1 |
||||||||||
|
Ren3Ren9 |
[64] |
V. vinifera cv. ‘Kismish vatkana’ 2 | |||||||||||
|
Ren6Ren7 |
[57] |
Yes |
Yes |
Yes |
Total |
[59] |
Ren1.2 |
V. vinifera cv. ‘Shavtsitka’ 3 |
Yes |
n.i. |
n.i. |
Partial |
[68] |
|
Ren2 |
V. cinerea2 |
Yes |
n.i. |
n.i. |
Partial |
||||||||
|
Ren3 |
‘Regent’ 4 |
Yes |
Yes |
Yes |
Partial |
||||||||
|
Ren4 |
V. romanetii2 |
Yes |
n.i. |
n.i. |
Partial |
[54] |
|||||||
|
Ren5 |
M. rotundifolia ‘Regale’ 1 |
n.i. |
n.i. |
n.i. |
Total |
[60] |
|||||||
|
Ren6 |
V. piasezki2 |
Yes |
n.i. |
n.i. |
Total |
[57] |
|||||||
|
Ren7 |
V. piasezki2 |
Yes |
n.i. |
n.i. |
Partial |
[57] |
|||||||
|
Ren8 |
Unknown 4 |
n.i. |
n.i. |
n.i. |
Partial |
[66] |
|||||||
|
Ren9 |
‘Regent’ 4 |
Yes |
n.i. |
n.i. |
Partial |
||||||||
|
Ren10 |
‘Seyval blanc’ 4 |
n.i. |
n.i |
n.i. |
Partial |
[67] |
|||||||
|
Ren11 |
Vitis aestivalis2 |
n.i. |
n.i. |
n.i |
Partial |
[61] |
5. Locus Stacking: The Search for Durable and Broad-Spectrum Resistance
Currently, one of the main objectives of grapevine breeding programs worldwide is the development of durable and strong resistance to powdery mildew, through independent modes of action. The most important desirable outcome of such programs is that the resistance must be durable. Because grapevine plants are productive for at least twenty years, resistance needs to be maintained through that period of time [55,57][55][57]. To achieve this goal, a pyramiding strategy has been proposed, which combines various resistance loci in the same genotype [109][86]. To ensure the durability of this resistance, it is necessary to mix loci that have different mechanisms of action, spectrums of target isolates, and contributions (minor and major) to the resistance [83,110][83][87]. Referring to this last aspect, it is important to consider that even though initially more promising results are observed when a gene or locus with a major effect is used, this can favor the selection of isolates of the fungus that are capable of overcoming this major resistance loci [110[87][88],111], and if resistance is based only on the presence of one gene, the fungus could mutate its effector and evade immune recognition [57]. A clear example of this is what happened between the Run1 locus and the Musc4 isolate, which is probably due to a long coexistence with M. rotundifolia, the donor specie of Run1, which likely mutated its effector to overcome the resistance conferred by this gene [51,85][51][85]. This response has not only been observed with Run1; Ren3 and Ren9 loci resistance were also overcome by a North American E. necator strain, despite these loci only conferring partial resistance [67]. These results suggest that in the case of the development of new grapevine cultivars with resistance to powdery mildew, it is important to consider the origin of the genes or loci when pyramiding, prioritizing the combination of resistance sources from species with diverse geographical origins. In the case of the development of resistant cultivars in North America, the high genetic variability of powdery mildew in that area [83] is a challenge for breeders. More studies are needed to evaluate the best combination of genes and loci for each viticultural area. Currently, the immune responses of some genotypes that have more than one source of resistance have already been characterized (Table 2). The presence of more than one resistant gene or loci does not generate a more intense resistance response in all the cases studied, demonstrating that combinations do not always generate additive effects (Table 2). This is the case of the Run1.2a/b genotypes that did not show any difference in PCD induction and secondary hyphae formation, compared to genotypes carrying just one of these loci [68]. Another example is the combination of Ren3 and Ren9, which did not generate an immune response that has an advantage in terms of the intensity or speed of the response compared to Ren3 alone [64]. This response has also been observed in Ren6Ren7 genotypes, which had an equal response to the Ren6 locus alone [57]. On the other hand, the combinations of Run1Run1.2a/b, Run1Ren1, and Run1Ren2 did show an additive effect, as the combination of both genes/loci generated a stronger immune response than the one triggered by each one individually. For example, the Run1Run1.2a/b genotypes showed less formation of secondary hyphae than each gene/locus separately [68], while in the case of Run1Ren1 genotypes, a more intense defense response was observed in terms of ROS production, callose accumulation, PCD, and activation of STILBENE SYNTHASE 36 (VvSTS36) and PENETRATION 1 (VvPEN1) than each of them separately [35]. The STS gene family encodes stilbene synthases, which catalyse the production of the stilbenes, compounds that have antimicrobial activities in plant defense [39]. PEN1 has a role in the traffic of secretory vesicles that could be associated with penetration resistance against powdery mildews [111][88]. For Run1Ren2 genotypes, a significant decrease in colony formation was seen compared to genotypes containing only Run1 or Ren2 [68].* Race-specific, as this effect was not seen with the Musc4 isolate.
6. Development of Genetic Resistance by Gene Editing
As an alternative approach to identify genes conferring resistance to E. necator, searching for susceptibility genes (S genes) can be an interesting strategy since inactivation of those S genes should lead to resistance to powdery mildew. An example of these S genes is the mildew locus O (MLO), which is conserved throughout the plant kingdom. Loss of function of certain members of the MLO gene family increases resistance to powdery mildew in A. thaliana, pea, tomato, wheat, and pepper. In Vitis, the combined silencing of VvMLO6, VvMLO7, and VvMLO11 produced a 77% decrease in E. necator infection [112][89]. However, although gene editing by Crispr–Cas9 of VvMLO3 did lead to an increase in resistance to powdery mildew, this was only observed in heterozygous plants, as the homozygous mutation produced plant death by necrosis, which suggests a pleiotropic function of this gene in Vitis [113][90].7. Final Remarks.
We are experiencing a devastated climate change phenomenon, which among its most important effects for food production are drought, the increase of insects and the elevated spread of pathogenic fungi. This scenario is especially worrying in woody plants such as vitis, because the study of its genome becomes very complex, due to the long waiting times for transformations, the genetic differences between each cultivar, and the high susceptibility of this plant to biotic and abiotic stress. Therefore, it becomes very important for current scientists to work to ensure the presence of this genus for future generations, in areas from biotechnology for genetic modifications, to engineering to create specific irrigation systems. But above all, take advantage of the knowledge that we have generated as a scientific community, disperse it in the population, and be able to achieve regulatory changes that allow us to dream, without irrational fears, in genetic improvements that benefit everyone.
References
- OIV. 2019 Statistical Report on World Vitiviniculture. Available online: https://www.oiv.int/public/medias/6782/oiv-2019-statistical-report-on-world-vitiviniculture.pdf (accessed on 12 December 2021).
- OIV. State of the World Viticultural Sector in 2020. Available online: https://www.oiv.int/public/medias/7909/oiv-state-of-the-world-vitivinicultural-sector-in-2020.pdf (accessed on 12 December 2021).
- Armijo, G.; Espinoza, C.; Loyola, R.; Restovic, F.; Santibáñez, C.; Schlechter, R.; Agurto, M.; Arce-Johnson, P. Grapevine biotechnology: Molecular approaches underlying abiotic and biotic stress responses. In Grape and Wine Biotechnology; Morata, A., Loira, I., Eds.; InTechOpen: Rijeka, Croatia, 2016; pp. 3–42. ISBN 978-953-51-2692-8.
- Feechan, A.; Kabbara, S.; Dry, I.B. Mechanisms of powdery mildew resistance in the Vitaceae family. Mol. Plant Pathol. 2011, 12, 263–274.
- Dry, I.B.; Feechan, A.; Anderson, C.; Jermakow, A.M.; Bouquet, A.; Adam-Blondon, A.-F.; Thomas, M.R. Molecular strategies to enhance the genetic resistance of grapevines to powdery mildew. Aust. J. Grape Wine Res. 2010, 16, 94–105.
- Pearson, R.C.; Gadoury, D.M. Grape powdery mildew. In Plant Diseases of International Importance; Kumar, J., Chaube, H.S., Singh, U.S., Mukhopadhyay, A.N., Eds.; Prentice Hall: Englewood Cliffs, NJ, USA, 1992; Volume III, pp. 129–146.
- Ferreira, R.B.; Monteiro, S.S.; Piçarra-Pereira, M.A.; Teixeira, A.R. Engineering grapevine for increased resistance to fungal pathogens without compromising wine stability. Trends Biotechnol. 2004, 22, 168–173.
- Pimentel, D.; Amaro, R.; Erban, A.; Mauri, N.; Soares, F.; Rego, C.; Martínez-Zapater, J.M.; Mithöfer, A.; Kopka, J.; Fortes, A.M. Transcriptional, hormonal, and metabolic changes in susceptible grape berries under powdery mildew Infection. J. Exp. Bot. 2021, 72, 6544–6569.
- Gaunt, R.E. The relationship between plant disease severity and yield. Annu. Rev. Phytopathol 1995, 33, 119–144.
- Madden, L.V.; Nutter, F.W. Modeling crop losses at the field scale. Can. J. Plant Pathol. 1995, 17, 124–137.
- Gadoury, D.M.; Cadle-Davidson, L.; Wilcox, W.F.; Dry, I.B.; Seem, R.C.; Milgroom, M.G. Grapevine powdery mildew (Erysiphe necator): A fascinating system for the study of the biology, ecology and epidemiology of an obligate biotroph: Grapevine powdery mildew. Mol. Plant Pathol. 2012, 13, 1–16.
- Calonnec, A.; Cartolaro, P.; Poupot, C.; Dubourdieu, D.; Darriet, P. Effects of Uncinula necator on the yield and quality of grapes (Vitis vinifera) and Wine. Plant Pathol. 2004, 53, 434–445.
- Lopez Pinar, A.; Rauhut, D.; Ruehl, E.; Buettner, A. Quantification of the changes in potent wine odorants as Induced by bunch rot (Botrytis cinerea) and powdery mildew (Erysiphe necator). Front. Chem. 2017, 5, 57.
- Amati, A.; Piva, A.; Castellari, M. Preliminary studies on the effect of Oidium tuckeri on the phenolic composition of grapes and wines. Vitis 1996, 35, 149–150.
- Marsh, E.; Alvarez, S.; Hicks, L.M.; Barbazuk, W.B.; Qiu, W.; Kovacs, L.; Schachtman, D. Changes in protein abundance during powdery mildew infection of leaf tissues of Cabernet Sauvignon grapevine (Vitis vinifera L.). Proteomics 2010, 10, 2057–2064.
- Fung, R.W.M.; Gonzalo, M.; Fekete, C.; Kovacs, L.G.; He, Y.; Marsh, E.; McIntyre, L.M.; Schachtman, D.P.; Qiu, W. Powdery mildew induces defense-oriented reprogramming of the transcriptome in a susceptible but not in a resistant grapevine. Plant Physiol. 2008, 146, 236–249.
- Saddhe, A.A.; Manuka, R.; Penna, S. Plant Sugars: Homeostasis and transport under abiotic stress in plants. Physiol. Plant. 2021, 171, 739–755.
- Stermer, B.A.; Bianchini, G.M.; Korth, K.L. Regulation of HMG-CoA reductase activity in plants. J. Lip. Res. 1994, 35, 1133–1140.
- Gadoury, D.M.; Seem, R.C.; Ficke, A.; Wilcox, W.F. Ontogenic resistance to powdery mildew in grape berries. Phytopathology 2003, 93, 547–555.
- Belpoggi, F.; Soffritti, M.; Guarino, M.; Lambertini, L.; Cevolani, D.; Maltoni, C. Results of Long-Term Experimental Studies on the Carcinogenicity of Ethylene-Bis-Dithiocarbamate (Mancozeb) in Rats. Ann. N. Y. Acad. Sci. 2006, 982, 123–136.
- Cecconi, S.; Paro, R.; Rossi, G.; Macchiarelli, G. The effects of the endocrine disruptors dithiocarbamates on the mammalian ovary with particular regard to mancozeb. CPD 2007, 13, 2989–3004.
- Calviello, G.; Piccioni, E.; Boninsegna, A.; Tedesco, B.; Maggiano, N.; Serini, S.; Wolf, F.I.; Palozza, P. DNA Damage and apoptosis induction by the pesticide mancozeb in rat cells: Involvement of the oxidative mechanism. Toxicol. Appl. Pharmacol. 2006, 211, 87–96.
- Mostafalou, S.; Abdollahi, M. Pesticides: An update of human exposure and toxicity. Arch. Toxicol. 2017, 91, 549–599.
- Pandey, A.; Jaiswar, S.; Ansari, N.; Deo, S.; Sankhwar, P.; Pant, S.; Upadhyay, S. Pesticide risk and recurrent pregnancy loss in Females of subhumid region of India. Niger Med. J. 2020, 61, 55.
- Komárek, M.; Čadková, E.; Chrastný, V.; Bordas, F.; Bollinger, J.C. Contamination of vineyard Soils with fungicides: A review of environmental and toxicological aspects. Environ. Int. 2010, 36, 138–151.
- Dumitriu, G.-D.; Teodosiu, C.; Cotea, V.V. Management of pesticides from vineyard to wines: Focus on wine safety and pesticides removal by emerging technologies. In Grapes and Wine; IntechOpen: London, UK, 2021; pp. 1–27.
- Peeters, A.; Lefebvre, O.; Balogh, L. A green deal for implementing agroecological systems: Reforming the common agricultural policy of the European Union. Landbauforsch. J. Sustain. Org. Agric. Syst. 2021, 70, 83–93.
- Borrello, M.; Cembalo, L.; Vecchio, R. Role of Information in consumers’ preferences for eco-sustainable genetic improvements in plant breeding. PLoS ONE 2021, 16, e0255130.
- Abramovitch, R.B.; Anderson, J.C.; Martin, G.B. Bacterial elicitation and evasion of plant innate immunity. Nat. Rev. Mol. Cell Biol 2006, 7, 601–611.
- Gao, Y.R.; Han, Y.-T.; Zhao, F.-L.; Li, Y.-J.; Cheng, Y.; Ding, Q.; Wang, Y.-J.; Wen, Y.-Q. Identification and utilization of a new Erysiphe necator isolate NAFU1 to quickly evaluate powdery mildew resistance in wild Chinese grapevine species using detached leaves. Plant Physiol. Biochem. 2016, 98, 12–24.
- He, P.; Shan, L.; Lin, N.-C.; Martin, G.B.; Kemmerling, B.; Nürnberger, T.; Sheen, J. Specific bacterial suppressors of MAMP signaling upstream of MAPKKK in Arabidopsis innate immunity. Cell 2006, 125, 563–575.
- Li, M.Y.; Jiao, Y.T.; Wang, Y.T.; Zhang, N.; Wang, B.B.; Liu, R.Q.; Yin, X.; Xu, Y.; Liu, G.T. CRISPR/Cas9-mediated VvPR4b editing decreases downy mildew resistance in grapevine (Vitis vinifera L.). Hortic. Res. 2020, 7, 149.
- Jones, J.D.G.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329.
- Qiu, W.; Feechan, A.; Dry, I. Current understanding of grapevine defense mechanisms against the biotrophic fungus (Erysiphe necator), the causal agent of powdery mildew disease. Hortic. Res. 2015, 2, 15020.
- Agurto, M.; Schlechter, R.O.; Armijo, G.; Solano, E.; Serrano, C.; Contreras, R.A.; Zúñiga, G.E.; Arce-Johnson, P. RUN1 and REN1 pyramiding in grapevine (Vitis vinifera cv. Crimson Seedless) displays an improved defense response leading to enhanced resistance to powdery mildew (Erysiphe necator). Front. Plant Sci. 2017, 8, 758.
- Dalla Costa, L.; Piazza, S.; Campa, M.; Flachowsky, H.; Hanke, M.-V.; Malnoy, M. Efficient heat-shock removal of the selectable marker gene in genetically modified grapevine. Plant Cell Tissue Organ Cult. 2016, 124, 471–481.
- Dalla Costa, L.; Piazza, S.; Pompili, V.; Salvagnin, U.; Cestaro, A.; Moffa, L.; Vittani, L.; Moser, C.; Malnoy, M. Strategies to produce T-DNA free CRISP red fruit trees via Agrobacterium tumefaciens stable gene transfer. Sci. Rep. 2020, 10, 20155.
- Malnoy, M.; Viola, R.; Jung, M.-H.; Koo, O.-J.; Kim, S.; Kim, J.-S.; Velasco, R.; Nagamangala Kanchiswamy, C. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 2016, 7, 1904.
- Dai, R.; Ge, H.; Howard, S.; Qiu, W. Transcriptional expression of stilbene synthase genes are regulated developmentally and differentially in response to powdery mildew in Norton and Cabernet Sauvignon grapevine. Plant Sci. 2012, 197, 70–76.
- Olivares, F.; Loyola, R.; Olmedo, B.; Miccono, M.d.l.Á.; Aguirre, C.; Vergara, R.; Riquelme, D.; Madrid, G.; Plantat, P.; Mora, R.; et al. CRISPR/Cas9 targeted editing of genes associated with fungal susceptibility in Vitis vinifera L. Cv. Thompson Seedless using Geminivirus-derived replicons. Front. Plant Sci. 2021, 12, 791030.
- Wildermut, M.C.; Dewdney, J.; Wu, G.; Ausubel, F.M. Isochorimate synthase is required to synthesize salicylic acid for plant defence. Nature 2001, 414, 562–565.
- Derksen, H.; Rampitsch, C.; Daayf, F. Signaling cross-talk in plant disease resistance. Plant Sci. 2013, 207, 79–87.
- Belhadj, A.; Saigne, C.; Telef, N.; Cluzet, S.; Bouscaut, J.; Corio-Costet, M.F.; Mérillon, J.-M. Methyl jasmonate induces defense responses in grapevine and triggers protection against Erysiphe necator. J. Agric. Food Chem. 2006, 54, 9119–9125.
- Belhadj, A.; Telef, N.; Cluzet, S.; Bouscaut, J.; Corio-Costet, M.-F.; Mérillon, J.-M. Ethephon elicits protection against Erysiphe necator in grapevine. J. Agric. Food Chem. 2008, 56, 5781–5787.
- Jacobs, A.L.; Dry, I.B.; Robinson, S.P. Induction of different pathogenesis-related cDNAs in grapevine infected with powdery mildew and treated with ethephon. Plant Pathol. 1999, 48, 325–336.
- Jiao, C.; Sun, X.; Yan, X.; Xu, X.; Yan, Q.; Gao, M.; Fei, Z.; Wang, X. Grape transcriptome response to powdery mildew infection: Comparative transcriptome profiling of Chinese wild grapes provides insights into powdery mildew resistance. Phytopathology 2021, 111, 2041–2051.
- Weng, K.; Li, Z.-Q.; Liu, R.-Q.; Wang, L.; Wang, Y.-J.; Xu, Y. Transcriptome of Erysiphe necator-infected Vitis pseudoreticulata leaves provides insight into grapevine resistance to powdery mildew. Hortic. Res. 2014, 1, 14049.
- Petrovic, T.; Perera, D.; Cozzolino, D.; Kravchuk, O.; Zanker, T.; Bennett, J.; Scott, E.S. Feasibility of discriminating powdery mildew-affected grape berries at harvest using mid-infrared attenuated total reflection spectroscopy and fatty acid profiling: Objective measures for grape powdery mildew. Australian J. Grape Wine Res. 2017, 23, 415–425.
- Vezzulli, S.; Dolzani, C.; Nicolini, D.; Bettinelli, P.; Migliaro, D.; Gratl, V.; Stedile, T.; Zatelli, A.; Dallaserra, M.; Clementi, S.; et al. Marker-assisted breeding for downy mildew, powdery mildew and phylloxera resistance at FEM. BIO Web Conf. 2019, 13, 01002.
- Grattapaglia, D.; Sederoff, R. Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: Mapping strategy and RAPD markers. Genetics 1994, 137, 1121–1137.
- Maul, H. Vitis International Variety Catalogue 2021. Available online: www.vivc.de (accessed on 15 December 2021).
- Barker, C.L.; Donald, T.; Pauquet, J.; Ratnaparkhe, M.B.; Bouquet, A.; Adam-Blondon, A.F.; Thomas, M.R.; Dry, I. Genetic and physical mapping of the grapevine powdery mildew resistance gene, Run1, using a bacterial artificial chromosome Library. Theor. Appl. Genet. 2005, 111, 370–377.
- Feechan, A.; Anderson, C.; Torregrosa, L.; Jermakow, A.; Mestre, P.; Wiedemann-Merdinoglu, S.; Merdinoglu, D.; Walker, A.R.; Cadle-Davidson, L.; Reisch, B.; et al. Genetic dissection of a TIR-NB-LRR locus from the wild North American grapevine species Muscadinia rotundifolia identifies paralogous genes conferring resistance to major fungal and oomycete pathogens in cultivated grapevine. Plant J. 2013, 76, 661–674.
- Ramming, D.W.; Gabler, F.; Smilanick, J.; Cadle-Davidson, M.; Barba, P.; Mahanil, S.; Cadle-Davidson, L. A single dominant locus, Ren4, confers rapid non-race-specific resistance to grapevine powdery mildew. Phytopathology 2011, 101, 502–508.
- Riaz, S.; Tenscher, A.C.; Ramming, D.W.; Walker, M.A. Using a limited mapping strategy to identify major QTLs for resistance to grapevine powdery mildew (Erysiphe necator) and their use in marker-assisted breeding. Theor. Appl. Genet. 2011, 122, 1059–1073.
- Mahanil, S.; Ramming, D.; Cadle-Davidson, M.; Owens, C.; Garris, A.; Myles, S.; Cadle-Davidson, L. Development of marker sets useful in the early selection of Ren4 powdery mildew resistance and seedlessness for table and raisin grape breeding. Theor. Appl. Genet. 2012, 124, 23–33.
- Pap, D.; Riaz, S.; Dry, I.B.; Jermakow, A.; Tenscher, A.C.; Cantu, D.; Oláh, R.; Walker, M.A. Identification of two novel powdery mildew resistance loci, Ren6 and Ren7, from the wild Chinese grape species Vitis piasezkii. BMC Plant Biol. 2016, 16, 170.
- Cadle-Davidson, L.E. A Perspective on Breeding and Implementing Durable Powdery Mildew Resistance. Acta Hortic. 2019, 541–548.
- Hoffmann, S.; Di Gaspero, G.; Kovács, L.; Howard, S.; Kiss, E.; Galbács, Z.; Testolin, R.; Kozma, P. Resistance to Erysiphe necator in the grapevine ‘Kishmish Vatkana’ is controlled by a single locus through restriction of hyphal growth. Theor. Appl. Genet. 2008, 116, 427–438.
- Blanc, S.; Wiedemann-Merdinoglu, S.; Dumas, V.; Mestre, P.; Merdinoglu, D. A reference genetic map of Muscadinia rotundifolia and identification of Ren5, a new major locus for resistance to grapevine powdery mildew. Theor. Appl. Genet. 2012, 125, 1663–1675.
- Karn, A.; Zou, C.; Brooks, S.; Fresnedo-Ramírez, J.; Gabler, F.; Sun, Q.; Ramming, D.; Naegele, R.; Ledbetter, C.; Cadle-Davidson, L. Discovery of the REN11 locus From Vitis aestivalis for stable resistance to grapevine powdery mildew in a family segregating for several unstable and tissue-specific quantitative resistance loci. Front. Plant Sci. 2021, 12, 733899.
- Ramming, D.W.; Gabler, F.; Smilanick, J.L.; Margosan, D.A.; Cadle-Davidson, M.; Barba, P.; Mahanil, S.; Frenkel, O.; Milgroom, M.G.; Cadle-Davidson, L. Identification of race-specific resistance in North American Vitis Spp. limiting Erysiphe Necator hyphal growth. Phytopathology 2012, 102, 83–93.
- Dalbó, M.A.; Ye, G.N.; Weeden, N.F.; Wilcox, W.F.; Reisch, B.I. Marker-assisted selection for powdery mildew resistance in grapes. J. Am. Soc. Hortic. Sci. 2001, 126, 83–89.
- Zendler, D.; Töpfer, R.; Zyprian, E. Confirmation and fine mapping of the resistance locus Ren9 from the grapevine cultivar ‘Regent’. Plants 2020, 10, 24.
- Zendler, D.; Schneider, P.; Töpfer, R.; Zyprian, E. Fine mapping of Ren3 reveals two loci mediating hypersensitive response against Erysiphe necator in grapevine. Euphytica 2017, 213, 68.
- Zyprian, E.; Ochßner, I.; Schwander, F.; Šimon, S.; Hausmann, L.; Bonow-Rex, M.; Moreno-Sanz, P.; Grando, M.S.; Wiedemann-Merdinoglu, S.; Merdinoglu, D.; et al. Quantitative trait loci affecting pathogen resistance and ripening of grapevines. Mol. Genet. Genomics 2016, 291, 1573–1594.
- Teh, S.L.; Fresnedo-Ramírez, J.; Clark, M.D.; Gadoury, D.M.; Sun, Q.; Cadle-Davidson, L.; Luby, J.J. Genetic dissection of powdery mildew resistance in interspecific half-sib grapevine families using SNP-based maps. Mol. Breeding 2017, 37, 1.
- Possamai, T.; Wiedemann-Merdinoglu, S.; Merdinoglu, D.; Migliaro, D.; De Mori, G.; Cipriani, G.; Velasco, R.; Testolin, R. Construction of a high-density genetic map and detection of a major QTL of resistance to powdery mildew (Erysiphe necator Sch.) in Caucasian grapes (Vitis vinifera L.). BMC Plant Biol. 2021, 21, 528.
- Feechan, A.; Kocsis, M.; Riaz, S.; Zhang, W.; Gadoury, D.M.; Walker, M.A.; Dry, I.B.; Reisch, B.; Cadle-Davidson, L. Strategies for RUN1 deployment using RUN2 and REN2 to manage grapevine powdery mildew informed by studies of race specificity. Phytopathology 2015, 105, 1104–1113.
- Welter, L.J.; Göktürk-Baydar, N.; Akkurt, M.; Maul, E.; Eibach, R.; Töpfer, R.; Zyprian, E.M. Genetic mapping and localization of quantitative trait loci affecting fungal disease resistance and leaf morphology in grapevine (Vitis vinifera L). Mol. Breeding 2007, 20, 359–374.
- Massonnet, M.; Vondras, A.M.; Cochetel, N.; Riaz, S.; Pap, D.; Minio, A.; Figueroa-Balderas, R.; Walker, M.A.; Cantu, D. Characterization of the grape powdery mildew genetic resistance loci in Muscadinia rotundifolia Trayshed. bioRxiv 2021.
- Yahiaoui, N.; Srichumpa, P.; Dudler, R.; Keller, B. Genome analysis at different ploidy levels allows cloning of the powdery mildew resistance gene Pm3b from hexaploid wheat. Plant J. 2004, 37, 528–538.
- Cao, A.; Xing, L.; Wang, X.; Yang, X.; Wang, W.; Sun, Y.; Qian, C.; Ni, J.; Chen, Y.; Liu, D.; et al. Serine/threonine kinase gene stpk-V, a key member of powdery mildew resistance gene Pm21, confes powdery mildew resistance in wheat. Proc. Natl. Acad. Sci. USA 2011, 108, 7727–7732.
- Hurni, S.; Brunner, S.; Buchmann, G.; Herren, G.; Jordan, T.; Krukowski, P.; Wicker, T.; Yahiaoui, N.; Mago, R.; Keller, B. Rye Pm8 and wheat Pm3 are orthologous genes and show evolutionary conservation of resistance function against powdery mildew. Plant J. 2013, 76, 957–969.
- He, H.; Zhu, S.; Jiang, Z.; Ji, Y.; Ji, J.; Qiu, D.; Li, H.; Bie, T. Pm21, encoding a typical CC-NBS-LRR protein, confers broad-spectrum resistance to wheat powdery mildew disease. Mol Plant. 2018, 11, 879–882.
- Zou, C.; Wang, H.; Li, Y.W.; Kong, Z.S.; Tang, D.S. The NB-LRR gene Pm60 confers powdery mildew resistance in wheat. New Phytol. 2018, 218, 298–309.
- Golzar, H.; Shankar, M.; D´Antuono, M. Responses of commercial wheat varieties and differential lines to western Australian powdery mildew (Blumeria graminis f. Sp. Tritici) populations. Australas. Plant Pathol. 2016, 45, 347–355.
- Xu, Q.; Wen, X.; Deng, X. Isolation of TIR and NonTIR NBS–LRR resistance gene analogues and identification of molecular markers linked to a powdery mildew resistance locus in Chestnut Rose (Rosa roxburghii Tratt). Theor. Appl. Genet. 2005, 111, 819–830.
- Thind, A.K.; Wicker, T.; Šimková, H.; Fossati, D.; Moullet, O.; Brabant, C.; Vrána, J.; Doležel, J.; Krattinger, S.G. Rapid cloning of genes in hexaploid wheat using cultivar-specific long-race chromosome assembly. Nat Biotechnol. 2017, 35, 793–796.
- Bouquet, A. Introduction Dans l’espèce Vitis vinifera L. d’un Caractère de Résistance à l’oidium (Uncinula necator Schw. Burr.) Issu de l’espèce Muscadinia rotundifolia (Michx.) Small. Vignevini 1986, 12, 141–146.
- Pauquet, J.; Bouquet, A.; This, P.; Adam-Blondon, A.F. Establishment of a local map of AFLP markers around the powdery mildew resistance gene Run1 in grapevine and assessment of their usefulness for marker assisted selection. Theor. Appl. Genet. 2001, 103, 1201–1210.
- Donald, T.M.; Pellerone, F.; Adam-Blondon, A.F.; Bouquet, A.; Thomas, M.R.; Dry, I.B. Identification of resistance gene analogs linked to a powdery mildew resistance locus in grapevine. Theor. Appl. Genet. 2002, 104, 610–618.
- Merdinoglu, D.; Schneider, C.; Prado, E.; Wiedemann-Merdinoglu, S.; Mestre, P. Breeding for durable resistance to downy and powdery mildew in grapevine. OENO One 2018, 52, 203–209.
- Anderson, C.; Choisne, N.; Adam-Blondon, A.-F.; Dry, I. Positional Cloning of Disease Resistance Genes in Grapevine. In Genetics, Genomics, and Breeding of Grapes; CRC Press: Boca Raton, FL, USA, 2016; pp. 214–238. ISBN 978-0-429-18941-8.
- Cadle-Davidson, L.; Mahanil, S.; Gadoury, D.M.; Kozma, P.; Reisch, B.I. Natural infection of Run1-positive vines by naïve genotypes of Erysiphe necator. Vitis 2011, 50, 85, 173–175.
- Mundt, C.C. Pyramiding for resistance durability: Theory and practice. Phytopathology 2018, 108, 792–802.
- McDonald, B.A.; Linde, C. Pathogen population genetics, evolutionary potential, and durable resistance. Annu. Rev. Phytopathol. 2002, 40, 349–379.
- Meyer, D.; Pajonk, S.; Micali, C.; O’Connell, R.; Schulze-Lefert, P. Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 2009, 57, 986–999.
- Peressotti, E.; Wiedemann-Merdinoglu, S.; Delmotte, F.; Bellin, D.; Di Gaspero, G.; Testolin, R.; Merdinoglu, D.; Mestre, P. Breakdown of resistance to grapevine downy mildew upon limited deployment of a resistant variety. BMC Plant Biol. 2010, 10, 147.
- Wan, D.-Y.; Guo, Y.; Cheng, Y.; Hu, Y.; Xiao, S.; Wang, Y.; Wen, Y.-Q. CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic. Res. 2020, 7, 116.
