The constant upward movement of data-driven medicine as a valuable option to enhance daily clinical practice has brought new challenges for data analysts to get access to valuable but sensitive data due to privacy considerations. One solution for most of these challenges are Distributed Analytics (DA) infrastructures, which are technologies fostering collaborations between healthcare institutions by establishing a privacy-preserving network for data sharing. However, in order to participate in such a network, a lot of technical and administrative prerequisites have to be made, which could pose bottlenecks and new obstacles for non-technical personnel during their deployment. Three major problems in the current state-of-the-art have been identified. Namely, the missing compliance with FAIR data principles, the automation of processes, and the installation.
- data profiling
- distributed analytics
- on-boarding
- Personal health Train
1. Introduction
2. Methods
2.1. FAIR Principles
The FAIR principles, firstly introduced by Wilkinson, M., Dumontier, M., Aalbersberg, I. et al., represent four basic guidelines for the improvement of scientific data management [16]. One driver of the FAIR data principles is the GO FAIR (https://www.go-fair.org/fair-principles/, accessed on 18 February 2022) initiative, which creates and coordinates so-called implementation networks (IN) to foster the establishment of these guidelines. As the acronym might suggest, these principles consist of the following four core pillars:-
Findable: To make data usage possible, researchers should be able to find digital assets. Each data object should have a persistent and unique identifier and should include rich metadata. Additionally, the metadata—or the identifier, respectively—should be stored in searchable resources.
-
Accessible: Open, free, and universal communication protocols should make data objects accessible by their identifier. Further, metadata should be archivable and available even when the corresponding data is not available.
-
Interoperable: Data should be interoperable with other data assets. This can be achieved by using formal, accessible, shared, and broadly applicable languages or vocabularies. Additionally, data should be referenceable from other data.
-
Reusable: To enable reusability, data should be equipped with usage licenses and detailed provenance and meet community standards.
2.2. Distributed Analytics
DA constitutes a paradigm shift in conventional data analysis [7][9][10][11][12][13][14][15]. At its core, approaches for DA perform data analysis decentrally. This means that analysis-ready data sets stay within institutional borders, and only the analysis code and its analysis results are transmitted between each institution. Finally, after analysing each data set, the aggregated analysis results return to the analyst, and from the obtained information, insights can be derived. The analysis can include basic statistics, queries, or even complex Machine Learning (ML) training routines. Besides the differences in the used technology, DA implementations can differ in their execution policies. There exists an incremental or a parallel execution of the analysis [13]. For the first approach, the involved data premises are set in succession, and the analysis is sent from one institution to the other. For the latter one, replicas of the analysis are sent to each institution and executed simultaneously. The results are sent back and aggregated centrally. Several prominent representatives of DA have been introduced during the past years, and basically, all of them follow these abstract analysis strategies [12][19][20][21][22]. One of such approaches is DataSHIELD (DS), which is client-server based [20][23]. DS uses a custom R-library to execute the analysis requests decentrally. DS has been applied to multiple use cases already (https://www.datashield.org/about/publications, accessed on 18 February 2022). Second, secure multi-party computation (SMPC) has attracted attention due to its security guarantees [21]. Instead of libraries, SMPC is protocol and encryption-based. Dependent on the analysis to be executed, these protocols could be more or less complex. The PHT is an infrastructure for privacy-preserving DA of patient-related health data. Figure 1 shows a high-level overview of the PHT. It consists of multiple components that can be roughly classified into centralised and client-side components. Clients—the so-called stations—are typically, but not necessarily, located in secured environments, i.e., behind a firewall at a specific institution which can be a research organization or even a company. Note that each institution can install as many stations as they want. The stations communicate with centralised components to retrieve analytical tasks, which are then executed on the client-side. Each station itself has access to the patient data, which is ideally already virtually integrated and homogenised. In Germany, patient data is currently homogenised based on the HL7 FHIR standard (https://www.hl7.org/fhir/, accessed on 18 February 2022) within the large Medical Informatics Initiative (https://www.medizininformatik-initiative.de/en/start, accessed on 18 February 2022). After an analysis ends, the station sends the results to the central components from which the next station can retrieve both the analysis algorithm (or better: script/program) and the intermediate results provided by previous stations.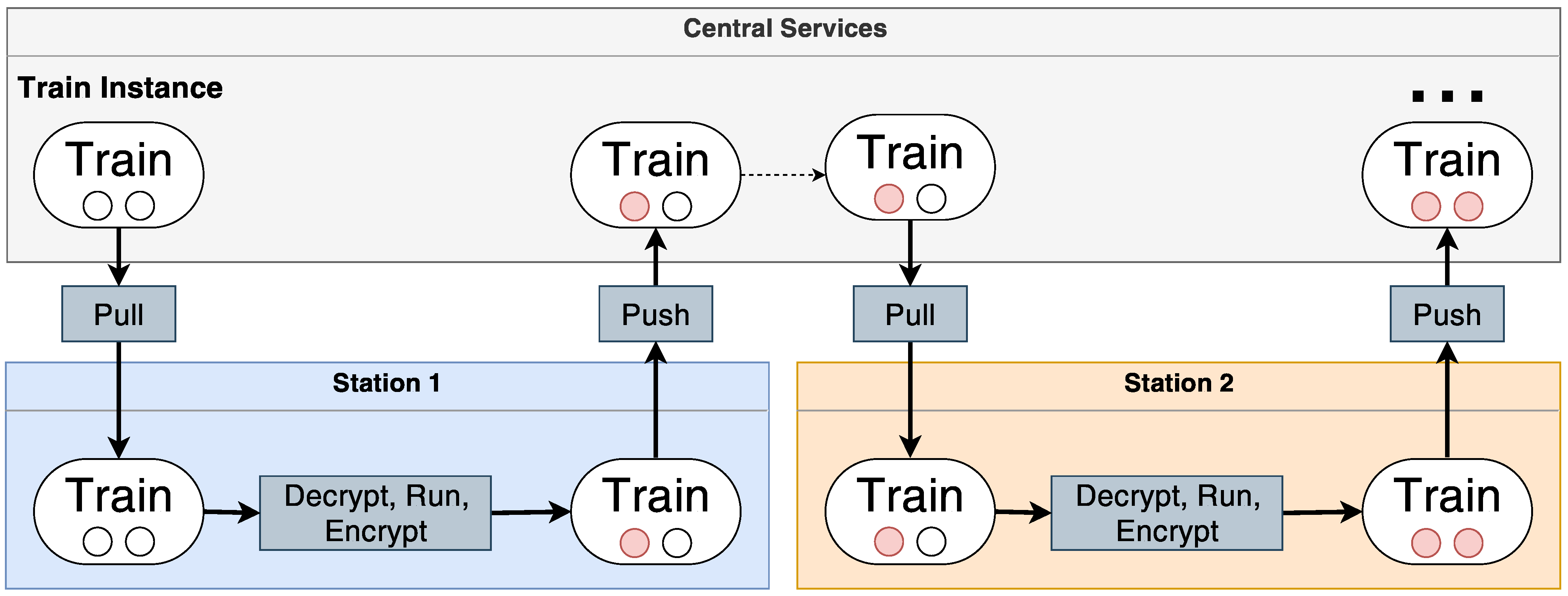
2.3. On-Boarding
To participate in a collaboration, one must set up all (technical) requirements to comply with the present conditions of this collaborative network. The mentioned collaborative network is the DA ecosystem, and the on-boarded object is the data-sharing institution. The term on-boarding is the process of providing all necessary installation materials and the installation itself. In order to on-board a so-called data computer in DS, researchers need an Opal server, which is open-source and online (https://opaldoc.obiba.org/en/latest/cookbook/r-datashield.html, accessed on 18 February 2022) available [20]. Gaye et al. state that the configuration of a DS does not require much IT expertise, and the installation can be conducted with no IT background [20]. Other possibilities (https://data2knowledge.atlassian.net/wiki/spaces/DSDEV/pages/1142325251/v6.1+Linux+Installation+Instructions, accessed on 18 February 2022) involve the deployment of a virtual machine hosting the DS functionalities and the manual input of IP addresses, which might pose challenges for non-technicians. Further, a connection to the DS client has to be established via REST over HTTPS, and the needed R libraries for DS applications have to be installed via a command-line interface (CLI) until the new station is ready for use [23]. The on-boarding process for a vantage6 station, a PHT-inspired technology, (so-called nodes) requires a priorly installed Docker daemon since vantage6 is a container-based infrastructure [22]. Moncada-Torres et al. state that the station administrator uses a CLI to start and configure the node’s core, which can be done using the well-established python package installer pip (https://docs.vantage6.ai/installation/node, accessed on 18 February 2022) [22]. Vantage6 further provides a CLI wizard for the node configuration (https://docs.vantage6.ai/usage/running-the-node/configuration, accessed on 18 February 2022), where all necessary information has to be provided by the user-such as server address, API key, or private key location for the encryption. Regarding the security protocol, the mandatory API key has to be exchanged between the server administrator and node manager. Vantage6 combines multiple nodes within one institution as organisation (https://docs.vantage6.ai/usage/preliminaries, accessed on 18 February 2022). According to their documentation, all nodes in the same organisation need to share the same private key. Therefore, also the private keys have to be exchanged separately. When the node starts, the corresponding public key of the private key is uploaded to the central server, which concludes the installation. Hewlett Packard (HP) has its own Swarm Learning (similar to DA) framework for the analysis of decentralised data [25]. Their on-boarding manual (https://github.com/HewlettPackard/swarm-learning/blob/master/docs/setup.md, accessed on 18 February 2022) states a sequence of Docker commands to be executed until the software is deployed. Lastly, a mandatory licence installation has to be conducted. In their whitepaper (https://www.hpe.com/psnow/doc/a50000344enw, accessed on 18 February 2022), they state that the on-boarding is an offline process and future participating parties need to communicate beforehand to find mutual requirements of the decentralised system. Another framework, called Flower, has been proposed by Beutel et al. [26]. They provide wrapper functions for the communication between each data node. Similar to vantage6, the necessary software can be downloaded using the pip installer (https://flower.dev, accessed on 18 February 2022). To connect clients with the server, the wrapper functions have to be implemented by the station admins such that a mutual encryption policy and a customisable communication configuration can be established. There are several potential shortcomings, which scholars have classified into three categories. First, some workflows do not contribute to FAIR data management or FAIRification of DA infrastructures as participating parties are not necessarily findable, for example. Therefore, each infrastructure acts as a blackbox to its users since the participating parties are not visible. Consequently, the connection information or other metadata for an institution has to be communicated through other channels to access the data. Additionally, there is a lack of automation in these workflows. Especially, the manual key exchange mechanism might pose some security risks if these are distributed within third-party channels. Further, the needed detailed configuration for some components (e.g., IP addresses, ports, certificates, secrets) might be another obstacle for non-technicians to set up a connection to the central services.3. On-Boarding Process for Distributed Analysis
3.1. Central Service
The central service (CS) component orchestrates the train images and performs the business logic. Each station has a dedicated repository for the trains such that each image can be pulled and pushed back after the execution. In the reference architecture, this repository is managed by an open-source container registry called Harbor (https://goharbor.io, accessed on 18 February 2022). To gain access to this repository, each station needs access credentials, which are provided by another component called Keycloak (https://www.keycloak.org, accessed on 18 February 2022)—an identity and access management (IAM) provider. Additionally, Vault (https://www.vaultproject.io, accessed on 18 February 2022) is used to securely store sensitive information and secrets such as the public keys of each station. Consequently, in order to participate in this infrastructure, it is required to distribute the Keycloak credentials and the Harbor repository connection information to each station. In return, the station has to send its public key to the CS such that it can be saved in the key store (Vault) for later usage.3.2. Station
The station software (client) is a fully-containerised application and can be accessed using a browser. Hence, a mandatory requirement is a Docker engine running on the host operating system. The installation of such an engine (https://www.docker.com/get-started, accessed on 18 February 2022) does not differ from a basic execution of a usual installer program, and therefore, no in-depth knowledge is needed. Essentially, the client software works as a remote control for the underlying Docker engine to execute the downloaded train images, which encapsulate the analysis code. To bring a station to life and set up the connection to the CS, it needs the connection credentials from the Keycloak instance and the Harbor repository address. In addition, it has to create a private/public key pair. The latter one has to be transmitted to the CS.3.3. On-Boarding Workflow
As the FAIR principles have suggested, to make a digital asset (in our case: the station) findable, it should have an identifier and should be findable in a searchable resource. The station registry is the leading component of the on-boarding process. It is a web-based application that hosts all available stations characteristics and their correspondence to the institution they belong to. In this way, it is similar to the Domain Name Service (DNS) of the internet and the authority for providing a list of available stations. The CS (and other software as well) can then reuse the information about available stations to let the scientist configure the route an analysis task should take. Therefore, the station registry is the place where new stations can be added as well as available stations are de-registered before they will be de-installed in their corresponding institutions. Scholars combine the action to register a new station with the on-boarding process. A status (online state) reflects whether a station is already available and can be included in a distributed analysis. While all users (including any software clients) can list available stations, only registered users can modify this list, i.e., adding new, deleting available and modifying characteristics (e.g., station name) of stations. To register a station, the station admin has to input basic information about the station, such as a responsible person, name, and contact information. Further, the station is assigned to an organisation or consortium, and one can select whether the station is publicly available or private (within the organisation). Scholars have carefully taken into consideration that the described data model is easily extendable. Finally, an on-boarding endpoint of a specific DA infrastructure can be selected to on-board the station to this ecosystem. Therefore, Scholars assume that each DA ecosystem provides an on-boarding interface, which can be triggered by the station registry. This further makes ourthe registry compatible with multiple ecosystems by simultaneously keeping all necessary information about the stations in one place.References
- Balicer, R.D.; Cohen-Stavi, C. Advancing Healthcare Through Data-Driven Medicine and Artificial Intelligence. In Healthcare and Artificial Intelligence; Nordlinger, B., Villani, C., Rus, D., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 9–15.
- Alyass, A.; Turcotte, M.; Meyre, D. From big data analysis to personalized medicine for all: Challenges and opportunities. BMC Med. Genom. 2015, 8, 33.
- Deo, R.C. Machine learning in medicine. Circulation 2015, 132, 1920–1930.
- Geifman, N.; Bollyky, J.; Bhattacharya, S.; Butte, A.J. Opening clinical trial data: Are the voluntary data-sharing portals enough? BMC Med. 2015, 13, 280.
- Sidey-Gibbons, J.A.M.; Sidey-Gibbons, C.J. Machine learning in medicine: A practical introduction. BMC Med. Res. Methodol. 2019, 19, 64.
- Giger, M.L. Machine Learning in Medical Imaging. J. Am. Coll. Radiol. 2018, 15, 512–520.
- Rieke, N.; Hancox, J.; Li, W.; Milletari, F.; Roth, H.; Albarqouni, S.; Bakas, S.; Galtier, M.N.; Landman, B.; Maier-Hein, K.; et al. The Future of Digital Health with Federated Learning. NPJ Digit. Med. 2020, 3, 119.
- Rosenblatt, M.; Jain, S.H.; Cahill, M. Sharing of Clinical Trial Data: Benefits, Risks, and Uniform Principles. Ann. Intern. Med. 2015, 162, 306–307.
- Sheller, M.J.; Reina, G.A.; Edwards, B.; Martin, J.; Bakas, S. Multi-Institutional Deep Learning Modeling Without Sharing Patient Data: A Feasibility Study on Brain Tumor Segmentation. In Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries. BrainLes (Workshop); Springer: Cham, Switzerland, 2019; Volume 11383, pp. 92–104.
- Sheller, M.J.; Edwards, B.; Reina, G.A.; Martin, J.; Pati, S.; Kotrotsou, A.; Milchenko, M.; Xu, W.; Marcus, D.; Colen, R.R.; et al. Federated learning in medicine: Facilitating multi-institutional collaborations without sharing patient data. Sci. Rep. 2020, 10, 12598.
- Welten, S.; Neumann, L.; Yediel, Y.U.; da Silva Santos, L.O.B.; Decker, S.; Beyan, O. DAMS: A Distributed Analytics Metadata Schema. Data Intell. 2021, 3, 528–547.
- Beyan, O.; Choudhury, A.; van Soest, J.; Kohlbacher, O.; Zimmermann, L.; Stenzhorn, H.; Karim, M.R.; Dumontier, M.; Decker, S.; da Silva Santos, L.O.B.; et al. Distributed Analytics on Sensitive Medical Data: The Personal Health Train. Data Intell. 2020, 2, 96–107.
- Chang, K.; Balachandar, N.; Lam, C.; Yi, D.; Brown, J.; Beers, A.; Rosen, B.; Rubin, D.L.; Kalpathy-Cramer, J. Distributed deep learning networks among institutions for medical imaging. J. Am. Med. Inform. Assoc. 2018, 25, 945–954.
- Shi, Z.; Zhovannik, I.; Traverso, A.; Dankers, F.J.W.M.; Deist, T.M.; Kalendralis, P.; Monshouwer, R.; Bussink, J.; Fijten, R.; Aerts, H.J.W.L.; et al. Distributed radiomics as a signature validation study using the Personal Health Train infrastructure. Sci. Data 2019, 6, 218.
- Deist, T.M.; Dankers, F.J.W.M.; Ojha, P.; Scott Marshall, M.; Janssen, T.; Faivre-Finn, C.; Masciocchi, C.; Valentini, V.; Wang, J.; Chen, J.; et al. Distributed learning on 20,000+ lung cancer patients—The Personal Health Train. Radiother. Oncol. 2020, 144, 189–200.
- Wilkinson, M.D.; Dumontier, M.; Aalbersberg, I.J.; Appleton, G.; Axton, M.; Baak, A.; Blomberg, N.; Boiten, J.W.; da Silva Santos, L.B.; Bourne, P.E.; et al. The FAIR Guiding Principles for scientific data management and stewardship. Sci. Data 2016, 3, 160018.
- Jacobsen, A.; Kaliyaperumal, R.; da Silva Santos, L.O.B.; Mons, B.; Schultes, E.; Roos, M.; Thompson, M. A Generic Workflow for the Data FAIRification Process. Data Intell. 2020, 2, 56–65.
- Sinaci, A.A.; Núñez-Benjumea, F.J.; Gencturk, M.; Jauer, M.L.; Deserno, T.; Chronaki, C.; Cangioli, G.; Cavero-Barca, C.; Rodríguez-Pérez, J.M.; Pérez-Pérez, M.M.; et al. From Raw Data to FAIR Data: The FAIRification Workflow for Health Research. Methods Inf. Med. 2020, 59, e21–e32.
- Welten, S.; Mou, Y.; Neumann, L.; Jaberansary, M.; Ucer, Y.Y.; Kirsten, T.; Decker, S.; Beyan, O. A Privacy-Preserving Distributed Analytics Platform for Health Care Data. Methods Inf. Med. 2022.
- Gaye, A.; Marcon, Y.; Isaeva, J.; LaFlamme, P.; Turner, A.; Jones, E.M.; Minion, J.; Boyd, A.W.; Newby, C.J.; Nuotio, M.L.; et al. DataSHIELD: Taking the analysis to the data, not the data to the analysis. Int. J. Epidemiol. 2014, 43, 1929–1944.
- Zhao, C.; Zhao, S.; Zhao, M.; Chen, Z.; Gao, C.Z.; Li, H.; An Tan, Y. Secure Multi-Party Computation: Theory, practice and applications. Inf. Sci. 2019, 476, 357–372.
- Moncada-Torres, A.; Martin, F.; Sieswerda, M.; Van Soest, J.; Geleijnse, G. VANTAGE6: An open source priVAcy preserviNg federaTed leArninG infrastructurE for Secure Insight eXchange. In Proceedings of the AMIA Annual Symposium, Online, 14–18 November 2020; American Medical Informatics Association: Bethesda, MD, USA, 2020; Volume 2020, pp. 870–877.
- Wilson, R.C.; Butters, O.W.; Avraam, D.; Baker, J.; Tedds, J.A.; Turner, A.; Murtagh, M.; Burton, P.R. DataSHIELD—New directions and dimensions. Data Sci. J. 2017, 16, 21.
- Mou, Y.; Welten, S.; Jaberansary, M.; Ucer Yediel, Y.; Kirsten, T.; Decker, S.; Beyan, O. Distributed Skin Lesion Analysis Across Decentralised Data Sources. Stud. Health Technol. Inform. 2021, 281, 352–356.
- Warnat-Herresthal, S.; Schultze, H.; Shastry, K.L.; Manamohan, S.; Mukherjee, S.; Garg, V.; Sarveswara, R.; Händler, K.; Pickkers, P.; Aziz, N.A.; et al. Swarm learning for decentralized and confidential clinical machine learning. Nature 2021, 594, 265–270.
- Beutel, D.J.; Topal, T.; Mathur, A.; Qiu, X.; Parcollet, T.; de Gusmão, P.P.; Lane, N.D. Flower: A friendly federated learning research framework. arXiv 2020, arXiv:2007.14390.
