Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Sonu Gandhi.
Salmonellosis is a major cause of foodborne infections, caused by Salmonella, posing a major health risk. It possesses the ability to infiltrate the food supply chain at any point throughout the manufacturing, distribution, processing or quality control process. Salmonella infection has increased severely and requires effective and efficient methods for early monitoring and detection.
- electrochemical biosensors
- Salmonella
1. Introduction
Salmonella is one of the most prevalent bacteria causing foodborne illnesses and death [1]. Salmonella is a Gram-negative, rod-shaped bacterium which belongs to the Enterobacteriaceae family [2,3][2][3]. Common standard methods for the identification of Salmonella bacteria are based on culture techniques, enzyme-linked immunosorbent assay (ELISA) and nucleic acid-based approaches [4]. Nucleic acid approaches include polymeric chain reaction (PCR), loop-mediated isothermal amplification (LAMP), nucleic acid sequence-based amplification (NASBA), micro arrays, recombinase polymerase amplification (RPA), and whole-genome sequencing (WGS). Some of these techniques require highly skilled individuals and sophisticated apparatus, and others consume a lot of time. As a result, the development of specific, sensitive and reliable technologies for quick detection is always needed for Salmonella diagnosis. Biosensors provide a number of advantages over laboratory-based assays, including increased sensitivity accuracy and specificity as well as the low cost, rapid response, in situ applications and potential for portability [5,6,7,8,9][5][6][7][8][9]. As a result, they have been identified as impressive alternatives for detecting Salmonella in food. There has been a surge in research investigations in this sector in recent years, with several publications. Numerous studies on this topic have also been published, with specific use of nanomaterials, electrochemical signal interpretation, and aptamer identification [10,11,12,13][10][11][12][13].
Electrochemical sensors detect the analyte of interest based on potentiometry, conductometry and impedimetric. Electrochemical sensing has been prevalent because of its rapid, specific detection response, sensitivity, ability to be miniaturized and integrated into point-of-care testing [14]. Furthermore, the utilization of different nanomaterials such as carbon nanotubes (CNTs), metallic nanoparticles, silica nanoparticles, metal oxide nanoparticles and organic nanoparticles enhances the detection limit in comparison with sensors with only molecular probes, antibodies or peptides [15]. Therefore, the selection of a bio-recognition element in combination with a nanomaterial is highly essential for electrochemical sensor development that would help with the sensitive and rapid detection of analytes [16].
The details of recently developed biosensors for sensing of Salmonella in food samples and the measures taken to miniaturize and integrate the sensors into point-of-care applications, including IOT-based (Temiz et al., 2015) [17], microfluidics (Rahmani et al., 2018) [18], CRISPR-based and potentiometric sensors (Figure 1).
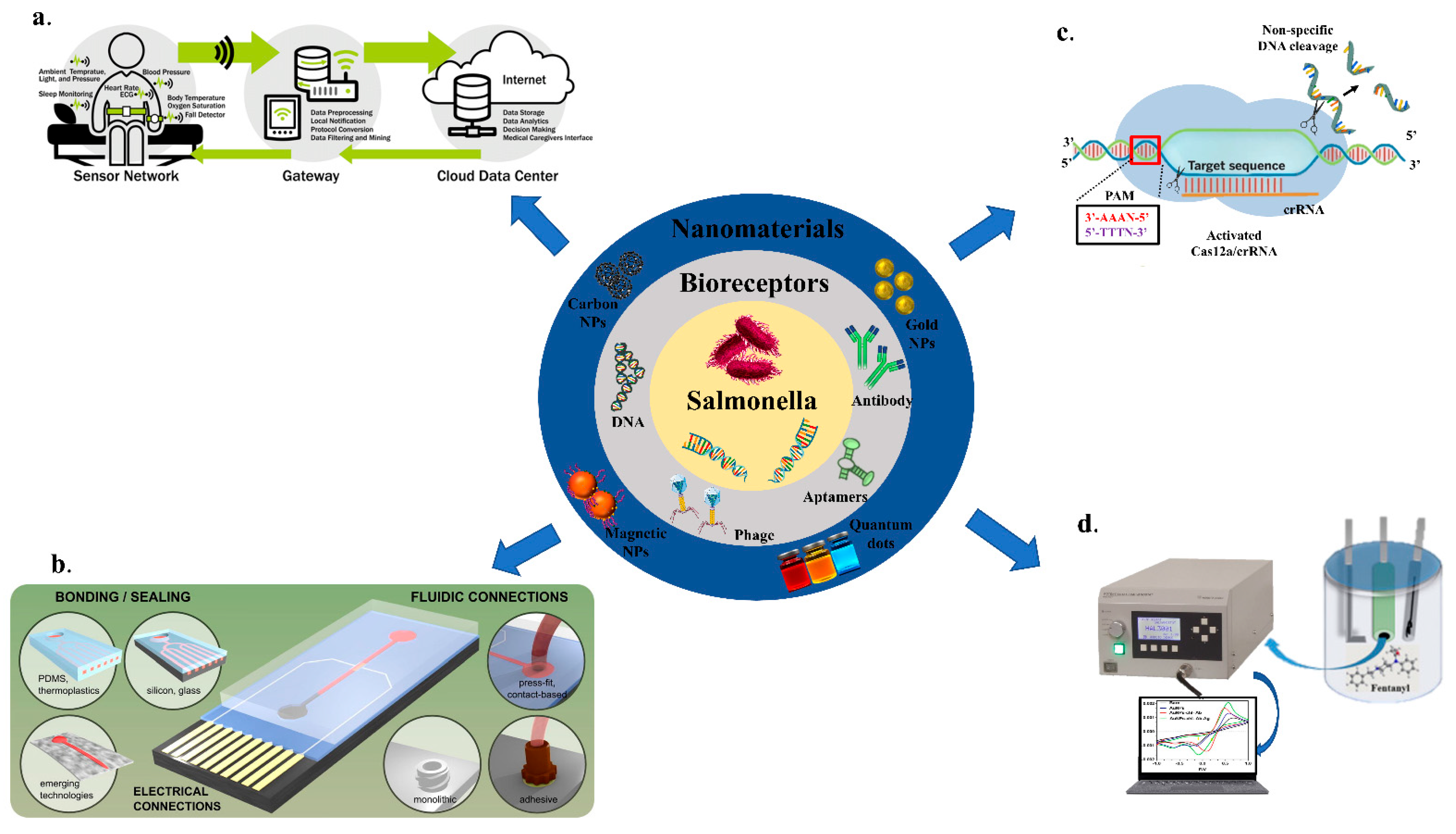
Figure 1. An overview of biosensors for the detection of Salmonella; (a) IOT-based; Adapted with permission from Ref. [17]. Copyright 2018, Elsevier. (b) Microfluidics based; Adapted with permission from Ref. [18]. Copyright 2015, Elsevier. (c) CRISPR/Cas-based; (d) Potentiometry-based biosensing.
2. Electrochemical Biosensors for Salmonella Detection
Sensors for the detection of Salmonella using the different bioreceptors, including various signal transducers processes, have increased their usage in recent years. Most of the sensors have been verified in food constituents, while others have the potential to be used in food samples. Different biosensing apparatuses for the detection of Salmonella have been addressed in this section, with a particular focus on electrochemical sensors. Their high sensitivity, low cost, and miniaturization potential make electrochemical biosensors the most commonly used sensing platform for the detection of Salmonella. Biosensors can be classified as amperometric, potentiometric, or conductometric based on the transducers used. A list of biosensors (electrochemical) for Salmonella detection strains are mentioned below in Table 1.
Table 1. Electrochemical biosensors for the detection of Salmonella strains.
| Sl No. | Serotype | Bio-Recognition Element | Detection Method | Range of Detection | LOD | References | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1. | Salmonella typhimurium | Aptamer | Potentiometry | 67–6.7 × 10 | 5 | CFU/mL | 10 CFU/mL | [71] | [19] | ||||
| 2. | Salmonella typhimurium | Aptamer | EIS | 10–10 | 8 | CFU/mL | 6 CFU/mL | [72] | [20] | ||||
| 3. | Salmonella typhimurium | Antibody | DPV | 10–10 | 7 | CFU/mL | 3 CFU/mL | [73] | [21] | ||||
| 4. | S. pullorum & S. gallinarum | Antibody | CV | 10 | 1 | –10 | 9 | CFU/mL | 1.61 × 10 | 1 | CFU/mL | [74] | [22] |
| 5. | S. enteritidis | Genosensor | SWAS V | 50 pg/mL–50 ng/mL | 0.5 ng/mL | [75] | [23] | ||||||
| 6. | S. pullorum | Antibody | EIS | 10 | 2 | to 10 | 6 | CFU /mL | 89 CFU /mL | [76] | [24] | ||
| 7. | S. pullorum and S. gallinarum | Antibody | CV | 10 | 4 | –10 | 9 | CFU/mL | 3.0 × 10 | 3 | CFU/ mL | [77] | [25] |
| 8. | Salmonella typhimurium | Antibody | EIS | 10–10 | 6 | CFU/mL | 10 CFU/mL | [78] | [26] | ||||
| 9. | Salmonella typhimurium | Aptamer | DPV | 20–20 | 7 | CFU/mL | 16 CFU/mL | [79] | [27] | ||||
| 10. | Salmonella enterica | Aptamer | DPV | 10–10 | 6 | CFU/mL | 10 CFU/mL | [80] | [28] | ||||
| 11. | Salmonella typhimurium | Antibody | DPV | 10–10 | 5 | CFU/ml | 5 CFU/mL | [81] | [29] |
2.1. Amperometric Biosensors for Salmonella Detection
Amperometry monitors variations in current when a constant voltage is applied for defined time. These sensors have acted as efficient models in the fast biosensing of Salmonella strains in food samples for decades, as shown in Figure 2 [82][30].
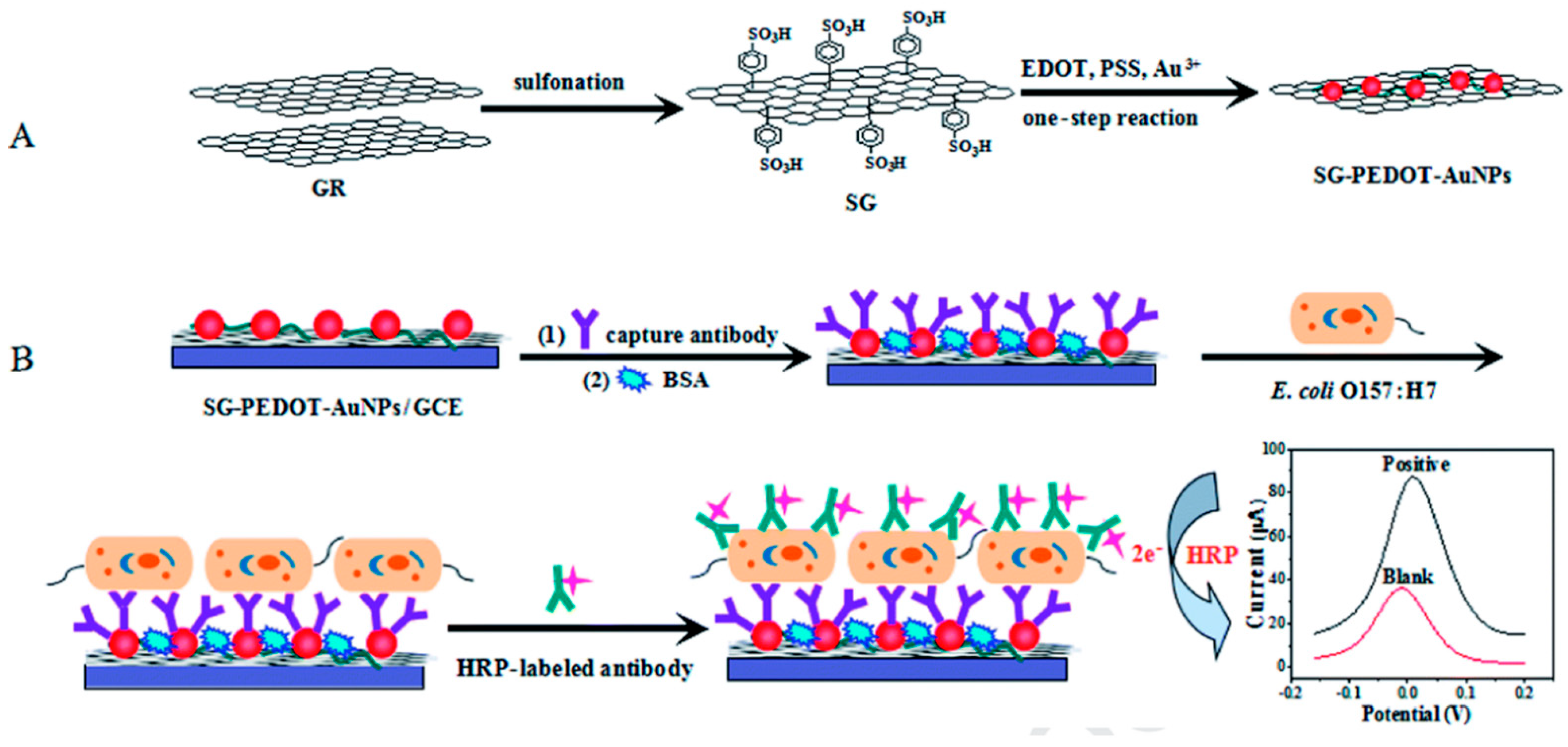
Figure 2. Principle of the sandwich assay. (A) Synthesis of sulfonated graphene (SG-PEDOT-AuNPs) composites and Poly-(3,4- 444 ethylenedioxythiophene) (PEDOT) in a single step reaction; (B) Schematic representation of electrochemical reaction where antibody was bound on the surface of glassy carbon electrode and detection of target by change in current. Adapted with permission from Ref. [82][30]. Copyright 2020, Elsevier.
Tyrosinase carbon paste electrode was used to measure phenol concentrations in a chicken carcass using an indirect method for detecting Salmonella typhimurium in 2.5 h with a LOD of 5 × 103 CFU/mL. The cells of Salmonella typhimurium were placed in between immunomagnetic beads (IMBs), and antibodies tagged with alkaline phosphatase (ALP). ALP was used as a catalyst for converting phenyl phosphate substrate to phenol, and tyrosinase carbon paste electrode used for the measurement. Furthermore, tyrosinase and horseradish peroxidase (HRP) electrode, which acted as a bio-enzyme, was used to enhance the LOD to 4.2 × 102 CFU/mL 5 [83][31]. In a study by Susana et al., Salmonella typhimurium could be detected in food samples using magneto immunoseparation with an LOD of 1 CFU/mL as shown in Figure 3 [84][32].
In contrast to such indirect detection principles, the following research on Salmonella detection centered on a direct sandwich ELISA approach. To precisely capture S. typhimurium, Tothill and Salam (2009) used the surface of a screen-printed gold (SPG) electrode to adhere the antibodies. Following the addition of HRP-labeled antibodies, a sandwich structure was created. Using the enzyme mediator/substrate system as 3,3′,5,5′-tetramethylbenzidine (TMB)/H2O2, this method was able to diagnose S. typhimurium cells at a concentration of around 21 CFU/mL [85][33].
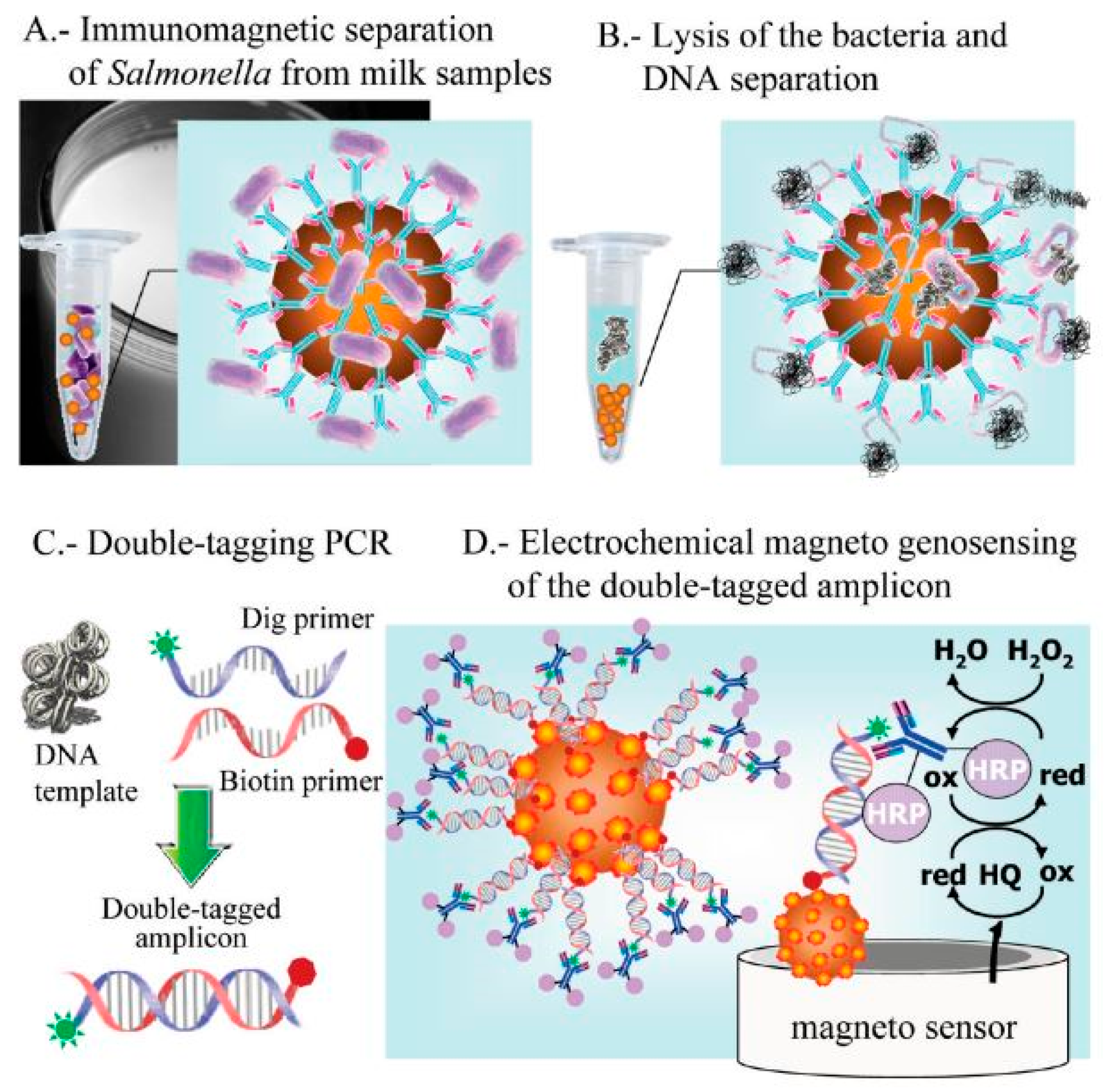
Figure 3. Electrochemical genosensing using immunomagnetic separation (IMS)/double-tagging PCR/m-GEC technique. (A) Separation of Salmonella contamination using Immunochromatographic separation in milk samples; (B) Bacterial lysis and separation of the nuclei acid; (C) Double-tagging of the nucleic acids; (D) Electrochemical genosensing (GEC) of double-tagged amplicon or the nucleic acid. Adapted with permission from Ref. [84][32]. Copyright 2009, American Chemical Society.
Later, Melo and colleagues (2018) doptimized several steps, including gold electrode pretreatment, antibody immobilization, enzyme–substrate, and concentrations of mediator with detection limit of 10 CFU/mL [86][34]. The amperometric method has been sensitive enough to detect Salmonella. Amperometric biosensors, on the other hand, rely on time-consuming labelling to boost the electrochemical response on the surface of electrode, limiting its use in the field.
2.2. Potentiometric Biosensors for Salmonella Detection
Under zero or insignificant current flow conditions, a high impedance voltmeter is used in potentiometric sensors for measurement of the change in electrical potential/electromotive force among the reference and working electrodes [87][35]. The cadmium ions amount produced during nanoparticle dissolution was used to measure the concentration of S. typhimurium with LOD of 20 CFU/mL in 75 min. The use of nanomaterials to generate/amplify the signal lengthens the time of analysis, and increases the assay’s complexity as shown in Figure 64 [88][36].
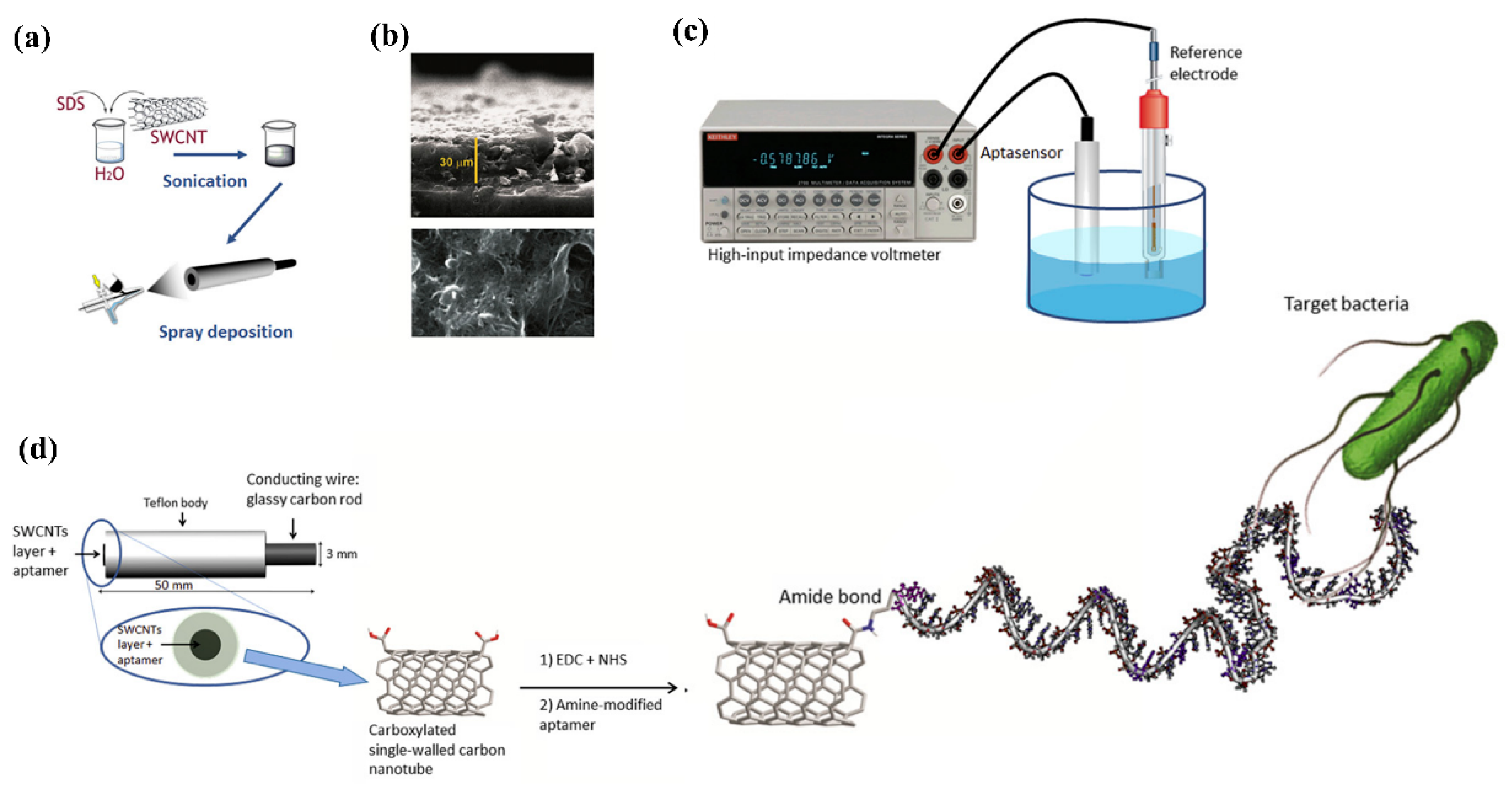
Figure 64. Fabrication of aptasensor on glassy carbon electrode (GCE). (a) Carbon nanotube deposition of on the surface of GCE; (b) Deposition of single-walled carbon nanotubes (SWCNTs) on GCE observed by (ESEM) environmental scanning electron microscope (representing a view from surface and lateral side). The surface view explains about the layer completely covering whole surface and the lateral view displays homogeneous height of the layer; (c) Potentiometric analysis; (d) Representation of an aptasensor for potentiometric analysis with specific aptamers functionalized with carboxylated SWCNTs and the correlation between the target bacteria and aptamer (Gustavo et al., 2013). Adapted with permission from Ref. [88][36]. Copyright 2013, Elsevier.
A label-free potentiometric detection was achieved in the presence of S. typhi with 0.2 CFU/mL in about 60 s. An important sensing technique of potentiometric sensors is based upon the surface blocking principle that has recently been presented for Salmonella detection, claiming to reach amplification capacities comparable to label-based techniques. The ion-flux internal marker was blocked via the detecting cell membrane attachment of Salmonella, resulting in a potentiometric response. Salmonella detection using these approaches helped in sensitive diagnosis with several CFU/mL of LODs within 1 h using transducers made up of paper-based strip electrode with a polymer inclusion membrane on an ion-selective electrode [89][37]. Exceptionally high sensitivity was obtained with the potentiometric biosensor. However, they have received less attention for Salmonella detection owing to the time-consuming tuning of experimental conditions and stabilization of the reference electrode.
2.3. Conductometric Biosensors for Salmonella Detection
Conductometry is a technique in which either no electrochemical reaction occurs at all on the electrodes or secondary reactions can be ignored. As a result, the conductivity of the electrolytic solution in the boundary layer is the most essential attribute in the conductometric approach, which varies according to a wide range of biological responses [90][38]. In comparison to other types of transducers, conductometric biosensors have several advantages such as the absence of the need for a reference electrode; operation at alternating voltage having a low amplitude, which helps prevent Faraday processes on electrodes; insensitivity to light, easy miniaturization, and integration using a low-cost thin-film standard technology [91][39]. First of all, they can be manufactured using low-cost thin-film standard technology. This, combined with the use of an improved approach for immobilizing biological material, leads to a significant reduction in both the primary cost of devices and the total cost of analyses. It is simple to use a differential measurement mode with inbuilt micro-biosensors, which compensates for external impacts and improves measurement accuracy significantly. The findings show that conductometric biosensors have a lot of promise [92][40]. Zarini et al. developed an electrochemical sandwich immunoassay and a reader was designed for signal measurement. The movement of bacteria was performed through electrical conductivity of the sensor with a detection limit of approximately 7.9 × 101 CFU/mL within a 10 min process [93][41]. As this is still a relatively new trend in the field of biosensors, the development of commercial devices for point-of-care applications has a bright future.
References
- Shivaprasad, H. Fowl typhoid and pullorum disease. Rev. Sci. Tech. l’OIE 2000, 19, 405–424.
- Silva, N.; Magalhães, J.M.; Freire, C.; Delerue-Matos, C. Electrochemical biosensors for Salmonella: State of the art and challenges in food safety assessment. Biosens. Bioelectron. 2018, 99, 667–682.
- Ansari, N.; Yazdian-Robati, R.; Shahdordizadeh, M.; Wang, Z.; Ghazvini, K. Aptasensors for quantitative detection of Salmonella Typhimurium. Anal. Biochem. 2017, 533, 18–25.
- Mahari, S.; Gandhi, S. Electrochemical immunosensor for detection of avian Salmonellosis based on electroactive reduced graphene oxide (rGO) modified electrode. Bioelectrochemistry 2021, 144, 108036.
- Roberts, A.; Mahari, S.; Shahdeo, D.; Gandhi, S. Label-free detection of SARS-CoV-2 Spike S1 antigen triggered by electroactive gold nanoparticles on antibody coated fluorine-doped tin oxide (FTO) electrode. Anal. Chim. Acta 2021, 1188, 339207.
- Shahdeo, D.; Gandhi, S. Next generation biosensors as a cancer diagnostic tool. In Biosensor Based Advanced Cancer Diagnostics; Academic Press: Cambridge, MA, USA, 2021; pp. 179–196.
- Roberts, A.; Chouhan, R.S.; Shahdeo, D.; Shrikrishna, N.S.; Kesarwani, V.; Horvat, M.; Gandhi, S. A Recent Update on Advanced Molecular Diagnostic Techniques for COVID-19 Pandemic: An Overview. Front. Immunol. 2021, 12, 5316.
- Kerry, R.G.; Mahapatra, G.P.; Maurya, G.K.; Patra, S.; Mahari, S.; Das, G.; Patra, J.K.; Sahoo, S. Molecular prospect of type-2 diabetes: Nanotechnology based diagnostics and therapeutic intervention. Rev. Endocr. Metab. Disord. 2020, 22, 421–451.
- Kujawska, M.; Bhardwaj, S.K.; Mishra, Y.K.; Kaushik, A. Using Graphene-Based Biosensors to Detect Dopamine for Efficient Parkinson’s Disease Diagnostics. Biosensors 2021, 11, 433.
- Ukhurebor, K.E.; Onyancha, R.B.; Aigbe, U.O.; Uk-Eghonghon, G.; Kerry, R.G.; Kusuma, H.S.; Darmokoesoemo, H.; Osibote, O.A.; Balogun, V.A. A Methodical Review on the Applications and Potentialities of Using Nanobiosensors for Disease Diagnosis. BioMed Res. Int. 2022, 2022, 1–20.
- Kerry, R.G.; Ukhurebor, K.E.; Kumari, S.; Maurya, G.K.; Patra, S.; Panigrahi, B.; Majhi, S.; Rout, J.R.; Rodriguez-Torres, M.D.P.; Das, G.; et al. A comprehensive review on the applications of nano-biosensor-based approaches for non-communicable and communicable disease detection. Biomater. Sci. 2021, 9, 3576–3602.
- Gandhi, S.; Banga, I.; Maurya, P.K.; Eremin, S.A. A gold nanoparticle-single-chain fragment variable antibody as an immunoprobe for rapid detection of morphine by dipstick. RSC Adv. 2018, 8, 1511–1518.
- Banga, I.; Tyagi, R.; Shahdeo, D.; Gandhi, S. Chapter 1—Biosensors and Their Application for the Detection of Avian Influenza Virus. In Nanotechnology in Modern Animal Biotechnology; Elsevier BV: Amsterdam, The Netherlands, 2019; pp. 1–16.
- Kaushik, A.; Khan, R.; Solanki, P.; Gandhi, S.; Gohel, H.; Mishra, Y.K. From Nanosystems to a Biosensing Prototype for an Efficient Diagnostic: A Special Issue in Honor of Professor Bansi D. Malhotra. Biosensors 2021, 11, 359.
- Naresh, V.; Lee, N. A Review on Biosensors and Recent Development of Nanostructured Materials-Enabled Biosensors. Sensors 2021, 21, 1109.
- Awang, M.S.; Bustami, Y.; Hamzah, H.H.; Zambry, N.S.; Najib, M.A.; Khalid, M.F.; Aziah, I.; Manaf, A.A. Advancement in Salmonella Detection Methods: From Conventional to Electrochemical-Based Sensing Detection. Biosensors 2021, 11, 346.
- Rahmani, A.M.; Gia, T.N.; Negash, B.S.; Anzanpour, A.; Azimi, I.; Jiang, M.; Liljeberg, P. Exploiting smart e-Health gateways at the edge of healthcare Internet-of-Things: A fog computing approach. Futur. Gener. Comput. Syst. 2018, 78, 641–658.
- Temiz, Y.; Lovchik, R.D.; Kaigala, G.V.; Delamarche, E. Lab-on-a-chip devices: How to close and plug the lab? Microelectron. Eng. 2015, 132, 156–175.
- Hasan, R.; Pulingam, T.; Appaturi, J.N.; Zifruddin, A.N.; Teh, S.J.; Lim, T.W.; Ibrahim, F.; Leo, B.F.; Thong, K.L. Carbon nanotube-based aptasensor for sensitive electrochemical detection of whole-cell Salmonella. Anal. Biochem. 2018, 554, 34–43.
- Bagheryan, Z.; Raoof, J.-B.; Golabi, M.; Turner, A.P.; Beni, V. Diazonium-based impedimetric aptasensor for the rapid label-free detection of Salmonella typhimurium in food sample. Biosens. Bioelectron. 2016, 80, 566–573.
- Bu, S.-J.; Wang, K.-Y.; Liu, X.; Ma, L.; Wei, H.-G.; Zhang, W.-G.; Liu, W.-S.; Wan, J.-Y. Ferrocene-functionalized nanocomposites as signal amplification probes for electrochemical immunoassay of Salmonella typhimurium. Mikrochim. Acta 2020, 187, 1–8.
- Wang, D.; Dou, W.; Chen, Y.; Zhao, G. Enzyme-functionalized electrochemical immunosensor based on electrochemically reduced graphene oxide and polyvinyl alcohol-polydimethylsiloxane for the detection of Salmonella pullorum & Salmonella gallinarum. RSC Adv. 2014, 4, 57733–57742.
- Zhang, D.; Huarng, M.C.; Alocilja, E.C. A multiplex nanoparticle-based bio-barcoded DNA sensor for the simultaneous detection of multiple pathogens. Biosens. Bioelectron. 2010, 26, 1736–1742.
- Fei, J.; Dou, W.; Zhao, G. Amperometric immunoassay for the detection of Salmonella pullorum using a screen - printed carbon electrode modified with gold nanoparticle-coated reduced graphene oxide and immunomagnetic beads. Mikrochim. Acta 2015, 183, 757–764.
- Fei, J.; Dou, W.; Zhao, G. A sandwich electrochemical immunosensor for Salmonella pullorum and Salmonella gallinarum based on a screen-printed carbon electrode modified with an ionic liquid and electrodeposited gold nanoparticles. Mikrochim. Acta 2015, 182, 2267–2275.
- Huang, F.; Xue, L.; Qi, W.; Cai, G.; Liu, Y.; Lin, J. An ultrasensitive impedance biosensor for Salmonella detection based on rotating high gradient magnetic separation and cascade reaction signal amplification. Biosens. Bioelectron. 2020, 176, 112921.
- Ge, C.; Yuan, R.; Yi, L.; Yang, J.; Zhang, H.; Li, L.; Nian, W.; Yi, G.; Ge, C.; Yuan, R.; et al. Target-induced aptamer displacement on gold nanoparticles and rolling circle amplification for ultrasensitive live Salmonella typhimurium electrochemical biosensing. J. Electroanal. Chem. 2018, 826, 174–180.
- Dinshaw, I.J.; Muniandy, S.; Teh, S.J.; Ibrahim, F.; Leo, B.F.; Thong, K.L. Development of an aptasensor using reduced graphene oxide chitosan complex to detect Salmonella. J. Electroanal. Chem. 2017, 806, 88–96.
- Xiang, C.; Li, R.; Adhikari, B.; She, Z.; Li, Y.; Kraatz, H.-B. Sensitive electrochemical detection of Salmonella with chitosan–gold nanoparticles composite film. Talanta 2015, 140, 122–127.
- Riu, J.; Giussani, B. Electrochemical biosensors for the detection of pathogenic bacteria in food. TrAC Trends Anal. Chem. 2020, 126, 115863.
- Fernández-Alba, A.R.; Guil, L.H.; López, G.D.; Chisti, Y. Toxicity of pesticides in wastewater: A comparative assessment of rapid bioassays. Anal. Chim. Acta 2001, 426, 289–301.
- Liébana, S.; Lermo, A.; Campoy, S.; Barbé, J.; Alegret, S.; Pividori, M.I. Magneto Immunoseparation of Pathogenic Bacteria and Electrochemical Magneto Genosensing of the Double-Tagged Amplicon. Anal. Chem. 2009, 81, 5812–5820.
- Salam, F.; Tothill, I.E. Detection of Salmonella typhimurium using an electrochemical immunosensor. Biosens. Bioelectron. 2009, 24, 2630–2636.
- Melo, A.M.A.; Alexandre, D.L.; Oliveira, M.R.F.; Furtado, R.F.; Borges, M.F.; Ribeiro, P.R.V.; Biswas, A.; Cheng, H.N.; Alves, C.R.; Figueiredo, E.A.T. Optimization and characterization of a biosensor assembly for detection of Salmonella Typhimurium. J. Solid State Electrochem. 2017, 22, 1321–1330.
- Velusamy, V.; Arshak, K.; Korostynska, O.; Oliwa, K.; Adley, C. An overview of foodborne pathogen detection: In the perspective of biosensors. Biotechnol. Adv. 2010, 28, 232–254.
- Zelada-Guillen, G.; Blondeau, P.; Rius, F.X.; Riu, J. Carbon nanotube-based aptasensors for the rapid and ultrasensitive detection of bacteria. Methods 2013, 63, 233–238.
- Silva, N.; Almeida, C.M.R.; Magalhães, J.M.; Gonçalves, M.P.; Freire, C.; Delerue-Matos, C. Development of a disposable paper-based potentiometric immunosensor for real-time detection of a foodborne pathogen. Biosens. Bioelectron. 2019, 141, 111317.
- Göpel, W.; Jones, T.A.; Kleitz, M.; Lundström, I.; Seiyama, T.; Hesse, J.; Zemel, J.N. Sensors, Chemical and Biochemical Sensors; John Wiley & Sons: Hoboken, NJ, USA, 1991; p. 734.
- Dzyadevych, S.; Jaffrezic-Renault, N. Conductometric biosensors. In Biological Identification; Woodhead Publishing: Cambridge, UK, 2014; pp. 153–193.
- Turner, A.; Karube, I.; Wilson, G.S. Biosensors: Fundamentals and Applications; Oxford University Press: Oxford, UK, 1987; p. 786.
- Muhammad-Tahir, Z.; Alocilja, E.C. A conductometric biosensor for biosecurity. Biosens. Bioelectron. 2003, 18, 813–819.
More
