Consideration of the totality of biological effects from cell death in multiple studies has led to the conceptualization of PANoptosis, a unique inflammatory cell death pathway that integrates components from other cell death pathways. PANoptosis is implicated in driving innate immune responses and inflammation and cannot be individually accounted for by pyroptosis, apoptosis, or necroptosis alone. PANoptosis is regulated by multifaceted macromolecular complexes called PANoptosomes.
- PANoptosis
- PANoptosome
- pyroptosis
- apoptosis
- necroptosis
- inflammatory cell death
- inflammasome
- inflammation
- innate immunity
- infection
- NLR
- caspase
- IRF1
- ZBP1
- RIPK1
- RIPK3
- MLKL
- NLRP3
- AIM2
- Pyrin
- caspase-1
- ASC
- caspase-8
- caspase-3
- caspase-7
- crosstalk
- plasticity
- re
1. Programmed Cell Death and PANoptosis
The innate immune system is the first line of defense against infection and cellular insults; innate immune receptors can recognize the molecular signatures of pathogens, called pathogen-associated molecular patterns (PAMPs), as well as components released by damaged cells, called damage-associated molecular patterns (DAMPs). The innate immune system activates genetically defined programmed cell death pathways in response to microbial infections or alterations in cellular homeostasis; among the most well characterized of these programmed cell death responses are pyroptosis, apoptosis, and necroptosis. Though canonically proposed as segregated cellular processes responding to individualized PAMPs and DAMPs, mounting evidence shows significant interactions between the components of pyroptosis, apoptosis, and necroptosis. Historically, the literature on cell death and innate immune signaling has used different terms to describe these interactions, such as ‘crosstalk’, ‘plasticity’, ‘redundancies’, and ‘molecular switches’. Consideration of the totality of biological effects from cell death in multiple studies has led to the conceptualization of PANoptosis [1,2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20], an inflammatory cell death pathway that integrates components from other cell death pathways. PANoptosis is implicated in driving innate immune responses and inflammation and cannot be individually accounted for by pyroptosis, apoptosis, or necroptosis alone. PANoptosis is regulated by PANoptosomes, multifaceted macromolecular complexes. Here, we review the key components of programmed cell death pathways and highlight the plasticity among pyroptosis, apoptosis, and necroptosis. We then discuss the conceptualization of PANoptosis, which continues to evolve over time based on data, and examine the current evidence supporting this concept.
The innate immune system is the first line of defense against infection and cellular insults; innate immune receptors can recognize the molecular signatures of pathogens, called pathogen-associated molecular patterns (PAMPs), as well as components released by damaged cells, called damage-associated molecular patterns (DAMPs). The innate immune system activates genetically defined programmed cell death pathways in response to microbial infections or alterations in cellular homeostasis; among the most well characterized of these programmed cell death responses are pyroptosis, apoptosis, and necroptosis. Though canonically proposed as segregated cellular processes responding to individualized PAMPs and DAMPs, mounting evidence shows significant interactions between the components of pyroptosis, apoptosis, and necroptosis. Historically, the literature on cell death and innate immune signaling has used different terms to describe these interactions, such as ‘crosstalk’, ‘plasticity’, ‘redundancies’, and ‘molecular switches’. Consideration of the totality of biological effects from cell death in multiple studies has led to the conceptualization of PANoptosis [1][2][3][4][5][6][7][8][9][10][11][12][13][14][15][16][17][18][19][20], an inflammatory cell death pathway that integrates components from other cell death pathways. PANoptosis is implicated in driving innate immune responses and inflammation and cannot be individually accounted for by pyroptosis, apoptosis, or necroptosis alone. PANoptosis is regulated by PANoptosomes, multifaceted macromolecular complexes. Here, researchers review the key components of programmed cell death pathways and highlight the plasticity among pyroptosis, apoptosis, and necroptosis. Researchers then discuss the conceptualization of PANoptosis, which continues to evolve over time based on data, and examine the current evidence supporting this concept.Key Components in Inflammatory Programmed Cell Death Pathways
Key Components in Inflammatory Programmed Cell Death Pathways
Among the most comprehensively studied cell death processes to date are pyroptosis, apoptosis, and necroptosis [21,22]. Each occurs in response to cellular insults, but they differ in terms of their molecular machinery. Pyroptosis is a lytic form of proinflammatory cell death that was originally described as a caspase-1-mediated death [23]. Pyroptotic cell death typically involves the formation of the inflammasome, a supramolecular platform that is composed of a sensor, adaptor protein ASC, and caspase-1 [24]. The five most well-known inflammasomes, which are named after their corresponding sensor based on the genetic characterization of sensors and triggers, are the NLR family inflammasomes, NLRP1 [24], NLRP3 [25,26,27], and NAIP/NLRC4 [28,29,30], as well as those formed by other sensors containing pyrin domains, such as Pyrin [31] and AIM2 [32,33]. Sensor activation polymerizes the adaptor protein ASC into prion-like structures referred to as ASC specks [34,35,36], which recruit caspase-1 to allow its autoproteolysis and activation [24,37]. Activated caspase-1 cleaves inflammatory cytokines IL-1β and IL-18 [38] as well as the pore-forming molecule gasdermin D (GSDMD) [39]. The GSDMD-mediated pores allow the release of the inflammatory cytokines [40,41,42,43,44] along with other inflammatory molecules such as HMGB1, which serve as DAMPs and further propagate an innate immune inflammatory response. GSDMD is also activated by caspase-11 (mice) and caspase-4/5 (humans) in the process of non-canonical inflammasome activation [39,40,41,44,45].
Apoptosis is a form of programmed cell death originally described as a ‘mechanism of controlled cell deletion’ characterized by its distinct morphological membrane blebbing and subsequent cell shrinkage [48]. It proceeds through either an extrinsic or intrinsic pathway, though both result in the activation of the same executioner caspases. The intrinsic pathway forms an APAF1-mediated apoptosome in response to homeostatic disruptions, such as DNA damage or loss of mitochondrial stability [49]. This multiprotein complex includes APAF1, cytochrome c, and the initiator caspase caspase-9 which, upon cleavage, activates the downstream effector/executioner caspases, caspase-3 and -7 [50,51]. Extrinsic apoptosis occurs after ligand binding to death receptors, such as Fas and TNF-α receptor (TNFR), on the cell surface; downstream of death receptor binding, FADD translocates to the receptor, which recruits caspase-8. Caspase-8 is the key extrinsic apoptotic initiator caspase which cleaves downstream caspases, caspase-3 and -7, to execute cell death [52,53]. Caspase-8 can also induce activation of intrinsic apoptosis by activating the proapoptotic molecule Bid [54,55,56], which translocates to the mitochondria to facilitate pore formation by BAX/BAK and induce mitochondrial outer membrane permeabilization (MOMP) and apoptosome formation [57,58].
Necroptosis, another lytic form of cell death, occurs in response to caspase-8 inhibition and is RIPK3- and MLKL-dependent [22,59,60,61]. The apoptotic caspase-8 typically blocks necroptosis by cleaving RIPK3, CYLD, and RIPK1 [62,63,64]. Necroptosis can be initiated in response to the activation of toll-like receptors (TLRs), death receptors, or through interferon (IFN) signaling [65]. A well-characterized necroptosis response is induced by TNF-α. Its binding to TNFR induces signaling that activates RIPK1 to become phosphorylated and, along with TRADD, FADD, and caspase-8, form complex II [66]. When caspase-8 is inhibited, RIPK1 interacts with RIPK3 to form a cell death-inducing necrosome. The RIPK1-RIPK3 complex promotes phosphorylation of MLKL, causing MLKL oligomerization. The MLKL multimer then translocates to the plasma membrane, where it interacts with phospholipids and forms pores [67,68]. In addition to MLKL phosphorylation by RIPK3, MLKL activity is also regulated by other post-translational modifications, such as ubiquitylation, which is necessary for higher-order oligomerization [69].
Within each of these cell death pathways, there are several regulators and variations in the signaling cascades that exist, making the complexity of these cellular processes, including regulatory components, cell- and trigger-specific responses, and time-dependent responses, limitless. Additional components and layers of complexity have been extensively reviewed elsewhere [22,78,79].
Within each of these cell death pathways, there are several regulators and auxiliary components. For example, additional inflammasome components have been identified that are involved in its regulation and activation, such as NEK7 [70][71][72][73] and DDX3X [74], as well as transcription factors such as IRF1 [75], IRF2 [46][47], and IRF8 [76]. Additionally, NINJ1 has been identified as a critical component for plasma membrane rupture [77]. There are also variations in the signaling cascades that exist within each programmed cell death pathway, making the complexity of these cellular processes, including regulatory components, cell- and trigger-specific responses, and time-dependent responses, limitless. Additional components and layers of complexity have been extensively reviewed elsewhere [22][78][79].2. Historical Development of the PANoptosis Concept
Understanding the activation and execution of inflammatory cell death pathways has been an active area of research, particularly given the clinical relevance of cell death pathways in infections, inflammatory diseases, cancers, and beyond [2,4,5,7,8,9,11,12,13,16,18,19,20]. As a result of these studies, several examples of crosstalk and flexibility have been identified between the molecular components of programmed cell death pathways. Here, we will limit our discussion to genetically defined examples over time.
Understanding the activation and execution of inflammatory cell death pathways has been an active area of research, particularly given the clinical relevance of cell death pathways in infections, inflammatory diseases, cancers, and beyond [2][4][5][7][8][9][11][12][13][16][18][19][20]. As a result of these studies, several examples of crosstalk and flexibility have been identified between the molecular components of programmed cell death pathways. Here, researchers will limit the discussion to genetically defined examples over time.At their core, apoptosis and necroptosis are intricately molecularly linked, given that TNF-induced caspase-8 activation drives apoptosis while inhibition of caspase-8 during this process drives necroptosis [80]. The rescue of caspase-8-deficient embryos by the loss of RIPK3 or MLKL has long been documented [81,82,83,84], and enzymatically active caspase-8 is critical in the regulation and balance of apoptosis and necroptosis [85,86].
At their core, apoptosis and necroptosis are intricately molecularly linked, given that TNF-induced caspase-8 activation drives apoptosis while inhibition of caspase-8 during this process drives necroptosis [80]. The rescue of caspase-8-deficient embryos by the loss of RIPK3 or MLKL has long been documented [81][82][83][84], and enzymatically active caspase-8 is critical in the regulation and balance of apoptosis and necroptosis [85][86].Beyond the intrinsic connection between apoptosis and necroptosis, caspase-1, an essential component of inflammasomes, cleaves apoptosis-associated caspase-7 during
Beyond the intrinsic connection between apoptosis and necroptosis, caspase-1, an essential component of inflammasomes, cleaves apoptosis-associated caspase-7 during Salmonella infection (NLRC4 inflammasome trigger) as well as in response to LPS + ATP stimulation (NLRP3 inflammasome trigger) [17]. The pyroptotic caspase-1 also cleaves apoptotic PARP1 in response to inflammasome-activating triggers [6], and loss of caspase-1 during Salmonella infection leads to activation of apoptotic proteins instead [87]. In addition, cells lacking pyroptotic caspase-1 and caspase-11 have reduced mitochondrial damage in response to inflammasome-activating triggers such as the NLRP3-activating LPS + ATP treatment or AIM2-activating dsDNA transfection [88], suggesting additional crosstalk between inflammasomes and apoptotic processes. Reciprocally, the apoptotic caspase-8 serves as a regulatory component of pyroptotic inflammasomes [19]. Fluorescence microscopy has shown the colocalization of caspase-8 and ASC in both pyroptosis-deficient and pyroptosis-sufficient cells in response to infections [12][89][90][91]. Additionally, caspase-8, along with FADD, is required to both prime and activate canonical (ligand-induced) and noncanonical (E. coli- or Citrobacter rodentium-induced) NLRP3 inflammasomes [19]. Caspase-8 can be recruited during NLRC4 and NLRP1b inflammasome formation [91][92][93] and at ASC specks involving multiple inflammasome sensors, such as NLRP3 and NLRC4 or AIM2 and Pyrin [12][94]; FADD can also be recruited to these ASC specks in response to FlaTox, a combination of the bacterial PAMPs Bacillus anthracis protective antigen and the N-terminus of lethal factor fused to Legionella pneumophila flagellin [93]. However, caspase-8 is not required forSalmonella infection (NLRC4 inflammasome trigger) as well as in response to LPS + ATP stimulation (NLRP3 inflammasome trigger) [17]. The pyroptotic caspase-1 also cleaves apoptotic PARP1 in response to inflammasome-activating triggers [6], and loss of caspase-1 during Salmonella infection leads to activation of apoptotic proteins instead [87]. In addition, cells lacking pyroptotic caspase-1 and caspase-11 have reduced mitochondrial damage in response to inflammasome-activating triggers such as the NLRP3-activating LPS + ATP treatment or AIM2-activating dsDNA transfection [88], suggesting additional crosstalk between inflammasomes and apoptotic processes. Reciprocally, the apoptotic caspase-8 serves as a regulatory component of pyroptotic inflammasomes [19]. Fluorescence microscopy has shown the colocalization of caspase-8 and ASC in both pyroptosis-deficient and pyroptosis-sufficient cells in response to infections [12,89,90,91]. Additionally, caspase-8, along with FADD, is required to both prime and activate canonical (ligand-induced) and noncanonical (E. coli- or Citrobacter rodentium-induced) NLRP3 inflammasomes [19]. Caspase-8 can be recruited during NLRC4 and NLRP1b inflammasome formation [91,92,93] and at ASC specks involving multiple inflammasome sensors, such as NLRP3 and NLRC4 or AIM2 and Pyrin [12,94]; FADD can also be recruited to these ASC specks in response to FlaTox, a combination of the bacterial PAMPs Bacillus anthracis protective antigen and the N-terminus of lethal factor fused to Legionella pneumophila flagellin [93]. However, caspase-8 is not required for Salmonella-induced cell death at 2, 6, and 24 h post-infection using an MOI of 1 or 10 [91], showcasing the variability of the roles of caspase-8 within the programmed cell death response.
-induced cell death at 2, 6, and 24 h post-infection using an MOI of 1 or 10 [91], showcasing the variability of the roles of caspase-8 within the programmed cell death response.Crosstalk has also been identified between cell death molecules by studying the totality of biological effects in disease processes. For example, inflammatory bone disease in mice carrying the Pstpip2
Crosstalk has also been identified between cell death molecules by studying the totality of biological effects in disease processes. For example, inflammatory bone disease in mice carrying the Pstpip2cmo mutation is driven by aberrant production of IL-1β and is strongly associated with inflammasome activation; however, inflammation persists despite deletion of caspase-1, NLRP3, or the combined deletion of caspase-8/RIPK3 (deletion of caspase-8 alone is embryonically lethal [84]). Inflammation is only rescued by the combined deletion of NLRP3 or caspase-1 with caspase-8/RIPK3 [16,18], highlighting the functional redundancies of pyroptotic molecules NLRP3 and caspase-1 with the apoptosis-necroptosis modulator caspase-8. In the context of infection, influenza A virus (IAV) induces activation of pyroptotic, apoptotic, and necroptotic proteins, and loss of RIPK3 protects against much of the cell death, but combined deletion of caspase-8 and RIPK3 is necessary to further reduce cell death [9], providing additional mechanistic evidence of overlaps in the functions of molecules involved in cell death activation.
mutation persists despite deletion of caspase-1 or combined deletion of caspase-8/RIPK3 (deletion of caspase-8 alone is embryonically lethal [84]); the inflammation is only rescued by the combined deletion of NLRP3 or caspase-1 with caspase-8/RIPK3 [16][18], highlighting the functional redundancies of pyroptotic molecules NLRP3 and caspase-1 with the apoptosis-necroptosis modulator caspase-8. In the context of infection, influenza A virus (IAV) induces activation of pyroptotic, apoptotic, and necroptotic proteins, and loss of RIPK3 protects against much of the cell death, but combined deletion of caspase-8 and RIPK3 is necessary to further reduce cell death [9], providing additional mechanistic evidence of overlaps in the functions of molecules involved in cell death activation.Beyond caspase-8 and RIPK3, the necroptotic molecule MLKL has also been implicated in crosstalk between cell death pathways. For example, ASC oligomerization to induce NLRP3 inflammasome activation can occur in response to treatment with TLR3 ligands and zVAD, but the ASC oligomerization is blocked in MLKL-deficient cells [95]. As oligomerized MLKL forms pores in the plasma membrane, a cascade of cellular consequences begins, including the efflux of potassium ions. This necroptosis-induced ionic efflux has been shown to activate the NLRP3 inflammasome [96,97]. Together, these data show how necroptosis and inflammasomes (pyroptosis) are interconnected.
Beyond caspase-8 and RIPK3, the necroptotic molecule MLKL has also been implicated in crosstalk between cell death pathways. For example, ASC oligomerization to induce NLRP3 inflammasome activation can occur in response to treatment with TLR3 ligands and zVAD, but the ASC oligomerization is blocked in MLKL-deficient cells [95]. As oligomerized MLKL forms pores in the plasma membrane, a cascade of cellular consequences begins, including the efflux of potassium ions. This necroptosis-induced ionic efflux has been shown to activate the NLRP3 inflammasome [96][97]. Together, these data show how necroptosis and inflammasomes (pyroptosis) are interconnected.Given the recently identified role of gasdermins in cell death, it has also been found that gasdermins mediate crosstalk between cell death pathways. GSDMD was initially identified as an executioner of pyroptotic cell death in response to caspase-1, caspase-4/-5 (human) or caspase-11 (mouse) cleavage [39,45]. Caspase-8 can also cleave GSDMD to activate pore formation and cell death during Yersinia infection [3,98,99,100]. Further studies have found that GSDMD can also be processed by the apoptosis-inducing caspase-3 in such a manner that renders GSDMD inactive, suppressing pyroptosis [101]. However, inflammasome and GSDMD activation in response to Shiga toxin 2 and LPS are also associated with increased mitochondrial ROS [102], and GSDMD can form pores in the mitochondrial membrane to release canonically pro-apoptotic molecules and activate caspase-3 in a BAK/BAX-independent manner [103,104]. Other members of the gasdermin family are also increasingly implicated in cell death crosstalk. Microarray and subsequent pathway analysis of inner ear samples from day 0 postnatal mice showed that the gene set involved in apoptosis is downregulated in mice lacking Gsdme as compared with wild-type controls [105]. Furthermore, GSDME can be cleaved by caspase-3, an apoptotic cell death effector, and can induce pyroptotic death [106,107]. In THP-1 cells lacking GSDMD, GSDME allows the release of IL-1β in response to nigericin, Val-boroPro, or Salmonella infection, though limited cell death was observed with endogenous GSDME expression levels in these cells [108]. In murine cells, NLRP3 inflammasome activation in GSDMD-deficient cells results in IL-1β and IL-18 release through caspase-8/-3 and GSDME activation [109]. GSDME serves in a feed-forward loop to promote caspase-3 activation by forming pores in the mitochondrial membrane and inducing the release of cytochrome c in response to traditional intrinsic and extrinsic apoptotic stimuli; overexpression studies have shown similar results with GSDMA [103]. Beyond these connections, in cells lacking pyroptosis via GSDMD-deficiency, caspase-1 can cleave caspase-3 and Bid to promote apoptotic cell death in response to inflammasome triggers such as LPS priming and poly(dA:dT) transfection, or during Salmonella infection [110,111]. Furthermore, the APAF1-apoptosome has been shown to interact with caspase-11 when cells are challenged with bile acid; the result is caspase-3 cleavage and the execution of pyroptotic death in a GSDME-mediated process [112]. Other pore-forming molecules may also be involved in this crosstalk, as pannexin-1 activation downstream of caspase-8 or -9 activation leads to NLRP3 inflammasome formation in a GSDMD- and GSDME-independent process [113].
Given the recently identified role of gasdermins in cell death, it has also been found that gasdermins mediate crosstalk between cell death pathways. GSDMD was initially identified as an executioner of pyroptotic cell death in response to caspase-1, caspase-4/-5 (human) or caspase-11 (mouse) cleavage [39][45]. Caspase-8 can also cleave GSDMD to activate pore formation and cell death during Yersinia infection [3][98][99][100]. Further studies have found that GSDMD can also be processed by the apoptosis-inducing caspase-3 in such a manner that renders GSDMD inactive, suppressing pyroptosis [101]. However, inflammasome and GSDMD activation in response to Shiga toxin 2 and LPS are also associated with increased mitochondrial ROS [102], and GSDMD can form pores in the mitochondrial membrane to release canonically proapoptotic molecules and activate caspase-3 in a BAK/BAX-independent manner [103][104]. Other members of the gasdermin family are also increasingly implicated in cell death crosstalk. Microarray and subsequent pathway analysis of inner ear samples from day-0 postnatal mice showed that the gene set involved in apoptosis is downregulated in mice lacking Gsdme as compared with wild-type controls [105]. Furthermore, GSDME can be cleaved by caspase-3, an apoptotic cell death effector, and can induce pyroptotic death [106][107]. In THP-1 cells lacking GSDMD, GSDME allows the release of IL-1β in response to nigericin, Val-boroPro, or Salmonella infection, though limited cell death was observed with endogenous GSDME expression levels in these cells [108]. In murine cells, NLRP3 inflammasome activation in GSDMD-deficient cells results in IL-1β and IL-18 release through caspase-8/-3 and GSDME activation [109]. GSDME serves in a feed-forward loop to promote caspase-3 activation by forming pores in the mitochondrial membrane and inducing the release of cytochrome c in response to traditional intrinsic and extrinsic apoptotic stimuli; overexpression studies have shown similar results with GSDMA [103]. Beyond these connections, in cells lacking pyroptosis via GSDMD-deficiency, caspase-1 can cleave caspase-3 and Bid to promote apoptotic cell death in response to inflammasome triggers such as LPS priming and poly(dA:dT) transfection, or during Salmonella infection [110][111]. Furthermore, the APAF1-apoptosome has been shown to interact with caspase-11 when cells are challenged with bile acid; the result is caspase-3 cleavage and the execution of pyroptotic death in a GSDME-mediated process [112]. Other pore-forming molecules may also be involved in this crosstalk, as pannexin-1 activation downstream of caspase-8 or -9 activation leads to NLRP3 inflammasome formation in a GSDMD- and GSDME-independent process [113].The overwhelming amount of evidence for the interconnectedness between cell death pathways has led to the conceptualization of PANoptosis as an inflammatory cell death pathway. The totality of biological effects in PANoptosis cannot be individually accounted for by pyroptosis, apoptosis, or necroptosis alone [2,3,4,5,6,7,8,9,10,11,12,13,14,15,16,17,18,19,20]. PANoptosis has been increasingly implicated in infectious and inflammatory diseases as well as in cancers and cancer therapies [2,3,4,5,7,8,9,10,11,12,13,14,15,16,18,20,114,115,116,117,118].
The overwhelming amount of evidence for the interconnectedness between cell death pathways has led to the conceptualization of PANoptosis as an inflammatory cell death pathway. The totality of biological effects in PANoptosis cannot be individually accounted for by pyroptosis, apoptosis, or necroptosis alone [2][3][4][5][6][7][8][9][10][11][12][13][14][15][16][17][18][19][20]. PANoptosis has been increasingly implicated in infectious and inflammatory diseases as well as in cancers and cancer therapies [2][3][4][5][7][8][9][10][11][12][13][14][15][16][18][20][114][115][116][117][1118].3. Prototypical Examples of PANoptosis
Here, wresearche will rs focus on the most mechanistically well-characterized examples of PANoptosis and PANoptosomes [3,7,9,10,11,12,20][3][7][9][10][11][12][20] (Figure 1).
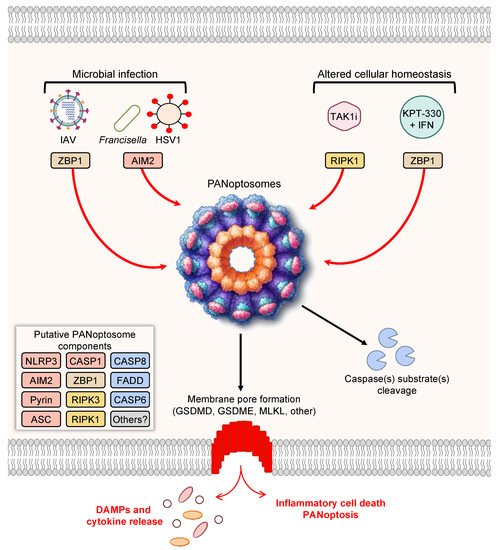
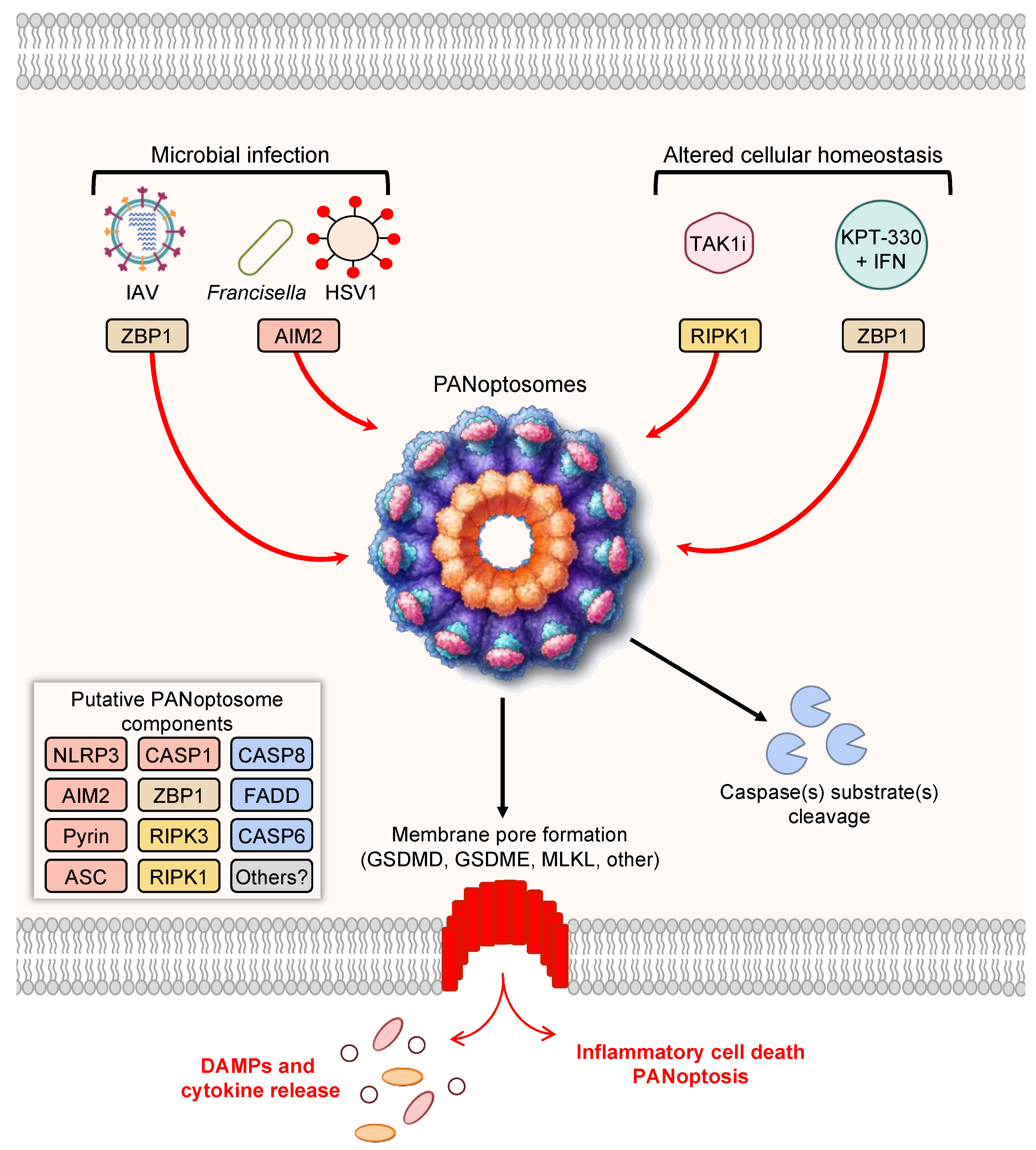
Figure 1. PANoptosis and PANoptosome formation. Upon exposure to cellular insults, such as microbial infection or altered cellular homeostasis, sensors can detect the perturbation and activate PANoptosis. Prototypical examples of PANoptosis are depicted here. Sensor activation can lead to the formation of a multiprotein complex, the PANoptosome. PANoptosomes have the potential to bring together diverse components from previously segregated cell death pathways. These may be dynamic complexes, and their protein composition may vary in trigger- and time-dependent manners. Potential PANoptosome components putatively include inflammasome sensors, such as nucleotide-binding oligomerization domain-like receptor family pyrin domain-containing 3 (NLRP3), absent in melanoma 2 (AIM2), Pyrin, Z-DNA-binding protein 1 (ZBP1), or others; apoptosis-associated speck-like protein containing a caspase activation and recruitment domain (ASC); caspase-1 (CASP1); receptor-interacting serine/threonine protein kinase 3 (RIPK3); RIPK1; caspase-8 (CASP8); Fas-associated protein with death domain (FADD); and/or caspase-6 (CASP6). PANoptosis involves membrane pore formation for the execution of cell death to release cytokines, such as IL-1β and IL-18, and DAMPs. Figure created with https://biorender.com/ (accessed on 17 March 2022).
As the conceptualization of PANoptosis implies, PANoptosis involves the activation of several molecules previously characterized as mediators of independent cell death pathways. For instance, Z-DNA-binding protein 1 (ZBP1) was previously known to induce necrosis in response to a mutant form of MCMV expressing a tetra-alanine RHIM substitution in vIRA (M45mutRHIM) [119], and ZBP1 interacts with the necroptotic molecules RIPK1 and RIPK3 [120]; however, more recent evidence has shown that ZBP1 acts as a cytosolic innate immune sensor for endogenous nucleic acids or during IAV infection to induce activation of the NLRP3 inflammasome, caspase-1, caspase-8, caspase-3, caspase-7 [7,9,11], and MLKL [7,11,121]. Molecularly, ZBP1 mediates the formation of a multiprotein ZBP1-PANoptosome complex, containing ZBP1, RIPK3, RIPK1, caspase-8, caspase-6, ASC, and NLRP3 [10,11]. To date, this complex has been characterized by immunoprecipitation [7,10,11], and immunofluorescence has also shown colocalization of caspase-8 and RIPK3 with ASC specks in individual cells during IAV infection [122]. Further studies, including biochemical analyses and cryo-EM evaluation, are needed to fully understand how components come together in individual cells. The ZBP1-PANoptosome complex can also be implicated in tumorigenesis, where ADAR1 acts as a negative regulator to prevent the interaction between ZBP1 and RIPK3 and promote tumorigenesis. Limiting the interaction between ADAR1 and ZBP1 by sequestering ADAR1 in the nucleus, through treatment with nuclear transport inhibitors (KPT-330) in conjunction with IFN, potentiates PANoptosis and limits tumorigenesis [7].
As the conceptualization of PANoptosis implies, PANoptosis involves the activation of several molecules previously characterized as mediators of independent cell death pathways. For instance, Z-DNA-binding protein 1 (ZBP1) was previously known to induce necrosis in response to a mutant form of MCMV expressing a tetra-alanine RHIM substitution in vIRA (M45mutRHIM) [119] and was shown to interact with the necroptotic molecules RIPK1 and RIPK3 [120], but more recent evidence has shown that ZBP1 acts as a cytosolic innate immune sensor for endogenous nucleic acids or during IAV infection to induce activation of the NLRP3 inflammasome, caspase-1, caspase-8, caspase-3, caspase-7 [7][9][11], and MLKL [7][11][121]. Molecularly, ZBP1 mediates the formation of a multiprotein ZBP1-PANoptosome complex, containing ZBP1, RIPK3, RIPK1, caspase-8, caspase-6, ASC, and NLRP3 [10][11]. To date, this complex has been characterized by immunoprecipitation [7][10][11], and immunofluorescence has also shown colocalization of caspase-8 and RIPK3 with ASC specks in individual cells during IAV infection [122]. Further studies, including biochemical analyses and cryo-EM evaluation, are needed to fully understand how components come together in individual cells. The ZBP1-PANoptosome complex can also be implicated in tumorigenesis, where ADAR1 acts as a negative regulator to prevent the interaction between ZBP1 and RIPK3 and promote tumorigenesis. Limiting the interaction between ADAR1 and ZBP1 by sequestering ADAR1 in the nucleus, through treatment with nuclear transport inhibitors (KPT-330) in conjunction with IFN, potentiates PANoptosis and limits tumorigenesis [7].Additionally, the inflammasome sensor AIM2 also initiates the formation of PANoptosome complexes during herpes simplex virus 1 (HSV1) and Francisella novicida infections. This PANoptosome, termed the AIM2-PANoptosome, contains AIM2, ZBP1, Pyrin, ASC, caspase-1, caspase-8, RIPK3, RIPK1, and FADD [12]. PANoptosomes have also been identified by immunoprecipitation during Yersinia infection, where RIPK1, RIPK3, caspase-8, FADD, ASC, and NLRP3 can be co-immunoprecipitated [3]. In the context of Yersinia infection, RIPK1 is necessary for activation of caspase-1, GSDMD, caspase-8, caspase-3, and caspase-7, but it negatively regulates the activation of MLKL, highlighting the multifaceted modulation of cell death effectors that can occur within PANoptosis [3]. PANoptosis has also been observed in response to TAK1 inhibition in macrophages, which can occur as a result of Yersinia infection due to its effector YopJ or in response to genetic mutations or treatment with TAK1 inhibitors [5,20,98,100]. In the case of TAK1-deficient macrophages, spontaneous PANoptosis occurs and is characterized by the activation of the NLRP3 inflammasome, caspase-1, caspase-3, caspase-8, and MLKL [5,20]; stimulation with LPS in TAK1-deficient macrophages induces colocalization of RIPK1, ASC, and caspase-8 in a RIPK1 kinase-independent manner [20].
Additionally, the inflammasome sensor AIM2 also initiates the formation of PANoptosome complexes during herpes simplex virus 1 (HSV1) and Francisella novicida infections. This PANoptosome, termed the AIM2-PANoptosome, contains AIM2, ZBP1, Pyrin, ASC, caspase-1, caspase-8, RIPK3, RIPK1, and FADD [12]. PANoptosomes have also been identified by immunoprecipitation during Yersinia infection, where RIPK1, RIPK3, caspase-8, FADD, ASC, and NLRP3 can be co-immunoprecipitated [3]. In the context of Yersinia infection, RIPK1 is necessary for activation of caspase-1, GSDMD, caspase-8, caspase-3, and caspase-7, but it negatively regulates the activation of MLKL, highlighting the multifaceted modulation of cell death effectors that can occur within PANoptosis [3]. PANoptosis has also been observed in response to TAK1 inhibition in macrophages, which can occur as a result of Yersinia infection due to its effector YopJ or in response to genetic mutations or treatment with TAK1 inhibitors [5][20][98][100]. In the case of TAK1-deficient macrophages, spontaneous PANoptosis occurs and is characterized by the activation of the NLRP3 inflammasome, caspase-1, caspase-3, caspase-8, and MLKL [5][20]; stimulation with LPS in TAK1-deficient macrophages induces colocalization of RIPK1, ASC, and caspase-8 in a RIPK1 kinase-independent manner [20].4. PANoptosis Regulation via IRF1
As with other cell death pathways, PANoptosis must be tightly regulated to control the execution of cell death. IRF1, a molecule long recognized for its roles in regulating cell death [123[123][124],124], is a key upstream regulator of PANoptosis. in manIn the absence of IRF1 during IAV infection, ZBP1 protein expression, along with NLRP3 inflammasome, caspase-1, caspase-8, caspase-3, and MLKL activation, are all reduced [75]. In the context of colorectal tumorigenesis, IRF1 facilitates the activation of PANoptosis to limit tumorigenesis [8]. PANoptosis has also been observed to be driven by IRF1 in response to the contembination of TNF and IFN-γ. TNF and IFN-γ release can occur physiologically during cytokine storm syndromes, including during SARS-CoV-2 infection [2], and together they induce PANoptosis through the JAK/IRF1 signaling axis [2][4]; this found toobservation has led to a mechanistic definition for cytokine storm as a life-threatening condition caused by excessive production of cytokines mediated by PANoptosis [125]. Addateitionally, the AIM2 inflammasome has also been shown to be regulated by IRF1 during Francisella infection [126], suggesting a possible regulatory role of IRF1 in PANoptosis mediated by the AIM2-PANoptosome, although this remains to be investigated. In the absence of IRF1 during IAV infection, ZBP1 protein expression, along with NLRP3 inflammasome, caspase-1, caspase-8, caspase-3, and MLKL activation, are all reduced [75]. In the context of colorectal tumorigenesis, IRF1 facilitates the activation of PANoptosis to limit tumorigenesis [8]. PANoptosis has also been observed to be driven by IRF1 in response to the combination of TNF and IFN-γ. TNF and IFN-γ release can occur physiologically during cytokine storm syndromes, including during SARS-CoV-2 infection [2], and together they induce PANoptosis through the JAK/IRF1 signaling axis [2,4]; this observation has led to a mechanistic definition for cytokine storm as a life-threatening condition caused by excessive production of cytokines mediated by PANoptosis [125]. Additionally, the AIM2 inflammasome has also been shown to be regulated by IRF1 during Francisella infection [126], suggesting a possible regulatory role of IRF1 in PANoptosis mediated by the AIM2-PANoptosome, although this remains to be investigated.5. Implications of PANoptosis in Disease
PANoptosis has been implicated across the disease spectrum, including in cerebral ischemia [114]; bacterial, viral, and fungal infections [3,9,10,11,12,13,14,15][3][9][10][11][12][13][14][115], including oral infections [115,116][115][116]; inflammatory diseases [2,5,16,18,20][2][5][16][18][20]; cancers [4,7,8][4][7][8]; and cancer therapies [4,7,117,118][4][7][117][118]. It will be important to improve ourthe understanding of this pathway and identify how previous descriptions of crosstalk, plasticity, redundancies, molecular switches, and interconnectedness among cell death processes fit within this inclusive concept to gain a holistic picture of cell death. Only when weresearchers identify all the ingredients in the PAN can weresearchers begin to effectively target these molecules and develop novel therapeutics to save lives and improve patient outcomes.
References
- Rajendra Karki; SangJoon Lee; Raghvendra Mall; Nagakannan Pandian; Yaqiu Wang; Bhesh Raj Sharma; Rk Subbarao Malireddi; Dong Yang; Sanja Trifkovic; Jacob A. Steele; et al.Jon P. ConnellyGella VishwanathMitnala SasikalaDuvvur Nageshwar ReddyPeter VogelShondra M. Pruett-MillerRichard WebbyColleen Beth JonssonThirumala-Devi Kanneganti ZBP1-dependent inflammatory cell death, PANoptosis, and cytokine storm disrupt IFN therapeutic efficacy during coronavirus infection. Science Immunology 2022, Online ahead of print, eabo6294, 10.1126/sciimmunol.abo6294.Malireddi, R.K.S.; Kesavardhana, S.; Kanneganti, T.D. ZBP1 and TAK1: Master Regulators of NLRP3 Inflammasome/Pyroptosis, Apoptosis, and Necroptosis (PAN-optosis). Front. Cell. Infect. Microbiol. 2019, 9, 406.
- Karki, R.; Sharma, B.R.; Tuladhar, S.; Williams, E.P.; Zalduondo, L.; Samir, P.; Zheng, M.; Sundaram, B.; Banoth, B.; Malireddi, R.K.S.; et al. Synergism of TNF-alpha and IFN-gamma Triggers Inflammatory Cell Death, Tissue Damage, and Mortality in SARS-CoV-2 Infection and Cytokine Shock Syndromes. Cell 2021, 184, 149–168.e117.
- Malireddi, R.K.S.; Kesavardhana, S.; Karki, R.; Kancharana, B.; Burton, A.R.; Kanneganti, T.D. RIPK1 Distinctly Regulates Yersinia-Induced Inflammatory Cell Death, PANoptosis. Immunohorizons 2020, 4, 789–796.
- Malireddi, R.K.S.; Karki, R.; Sundaram, B.; Kancharana, B.; Lee, S.; Samir, P.; Kanneganti, T.D. Inflammatory Cell Death, PANoptosis, Mediated by Cytokines in Diverse Cancer Lineages Inhibits Tumor Growth. Immunohorizons 2021, 5, 568–580.
- Malireddi, R.K.S.; Gurung, P.; Mavuluri, J.; Dasari, T.K.; Klco, J.M.; Chi, H.; Kanneganti, T.D. TAK1 restricts spontaneous NLRP3 activation and cell death to control myeloid proliferation. J. Exp. Med. 2018, 215, 1023–1034.
- Malireddi, R.K.; Ippagunta, S.; Lamkanfi, M.; Kanneganti, T.D. Cutting edge: Proteolytic inactivation of poly(ADP-ribose) polymerase 1 by the Nlrp3 and Nlrc4 inflammasomes. J. Immunol. 2010, 185, 3127–3130.
- Karki, R.; Sundaram, B.; Sharma, B.R.; Lee, S.; Malireddi, R.K.S.; Nguyen, L.N.; Christgen, S.; Zheng, M.; Wang, Y.; Samir, P.; et al. ADAR1 restricts ZBP1-mediated immune response and PANoptosis to promote tumorigenesis. Cell Rep. 2021, 37, 109858.
- Karki, R.; Sharma, B.R.; Lee, E.; Banoth, B.; Malireddi, R.K.S.; Samir, P.; Tuladhar, S.; Mummareddy, H.; Burton, A.R.; Vogel, P.; et al. Interferon regulatory factor 1 regulates PANoptosis to prevent colorectal cancer. JCI Insight 2020, 5, e136720.
- Kuriakose, T.; Man, S.M.; Subbarao Malireddi, R.K.; Karki, R.; Kesavardhana, S.; Place, D.E.; Neale, G.; Vogel, P.; Kanneganti, T.D. ZBP1/DAI is an innate sensor of influenza virus triggering the NLRP3 inflammasome and programmed cell death pathways. Sci. Immunol. 2016, 1, aag2045.
- Christgen, S.; Zheng, M.; Kesavardhana, S.; Karki, R.; Malireddi, R.K.S.; Banoth, B.; Place, D.E.; Briard, B.; Sharma, B.R.; Tuladhar, S.; et al. Identification of the PANoptosome: A Molecular Platform Triggering Pyroptosis, Apoptosis, and Necroptosis (PANoptosis). Front. Cell. Infect. Microbiol. 2020, 10, 237.
- Zheng, M.; Karki, R.; Vogel, P.; Kanneganti, T.D. Caspase-6 is a key regulator of innate immunity, inflammasome activation and host defense. Cell 2020, 181, 674–687.e13.
- Lee, S.; Karki, R.; Wang, Y.; Nguyen, L.N.; Kalathur, R.C.; Kanneganti, T.D. AIM2 forms a complex with pyrin and ZBP1 to drive PANoptosis and host defence. Nature 2021, 597, 415–419.
- Kesavardhana, S.; Malireddi, R.K.S.; Burton, A.R.; Porter, S.N.; Vogel, P.; Pruett-Miller, S.M.; Kanneganti, T.D. The Zα2 domain of ZBP1 is a molecular switch regulating influenza-induced PANoptosis and perinatal lethality during development. J. Biol. Chem. 2020, 295, 8325–8330.
- Banoth, B.; Tuladhar, S.; Karki, R.; Sharma, B.R.; Briard, B.; Kesavardhana, S.; Burton, A.; Kanneganti, T.D. ZBP1 promotes fungi-induced inflammasome activation and pyroptosis, apoptosis, and necroptosis (PANoptosis). J. Biol. Chem. 2020, 295, 18276–18283.
- Zheng, M.; Williams, E.P.; Malireddi, R.K.S.; Karki, R.; Banoth, B.; Burton, A.; Webby, R.; Channappanavar, R.; Jonsson, C.B.; Kanneganti, T.D. Impaired NLRP3 inflammasome activation/pyroptosis leads to robust inflammatory cell death via caspase-8/RIPK3 during coronavirus infection. J. Biol. Chem. 2020, 295, 14040–14052.
- Lukens, J.R.; Gurung, P.; Vogel, P.; Johnson, G.R.; Carter, R.A.; McGoldrick, D.J.; Bandi, S.R.; Calabrese, C.R.; Walle, L.V.; Lamkanfi, M.; et al. Dietary modulation of the microbiome affects autoinflammatory disease. Nature 2014, 516, 246–249.
- Lamkanfi, M.; Kanneganti, T.D.; Van Damme, P.; Vanden Berghe, T.; Vanoverberghe, I.; Vandekerckhove, J.; Vandenabeele, P.; Gevaert, K.; Nunez, G. Targeted peptidecentric proteomics reveals caspase-7 as a substrate of the caspase-1 inflammasomes. Mol. Cell. Proteom. 2008, 7, 2350–2363.
- Gurung, P.; Burton, A.; Kanneganti, T.D. NLRP3 inflammasome plays a redundant role with caspase 8 to promote IL-1β–mediated osteomyelitis. Proc. Natl. Acad. Sci. USA 2016, 113, 4452–4457.
- Gurung, P.; Anand, P.K.; Malireddi, R.K.; Vande Walle, L.; Van Opdenbosch, N.; Dillon, C.P.; Weinlich, R.; Green, D.R.; Lamkanfi, M.; Kanneganti, T.D. FADD and caspase-8 mediate priming and activation of the canonical and noncanonical Nlrp3 inflammasomes. J. Immunol. 2014, 192, 1835–1846.
- Malireddi, R.K.S.; Gurung, P.; Kesavardhana, S.; Samir, P.; Burton, A.; Mummareddy, H.; Vogel, P.; Pelletier, S.; Burgula, S.; Kanneganti, T.D. Innate immune priming in the absence of TAK1 drives RIPK1 kinase activity-independent pyroptosis, apoptosis, necroptosis, and inflammatory disease. J. Exp. Med. 2020, 217, e20191644.
- Kesavardhana, S.; Malireddi, R.K.S.; Kanneganti, T.D. Caspases in Cell Death, Inflammation, and Pyroptosis. Annu. Rev. Immunol. 2020, 38, 567–595.
- Galluzzi, L.; Vitale, I.; Aaronson, S.A.; Abrams, J.M.; Adam, D.; Agostinis, P.; Alnemri, E.S.; Altucci, L.; Amelio, I.; Andrews, D.W.; et al. Molecular mechanisms of cell death: Recommendations of the Nomenclature Committee on Cell Death 2018. Cell Death Differ. 2018, 25, 486–541.
- Cookson, B.T.; Brennan, M.A. Pro-inflammatory programmed cell death. Trends Microbiol. 2001, 9, 113–114.
- Martinon, F.; Burns, K.; Tschopp, J. The inflammasome: A molecular platform triggering activation of inflammatory caspases and processing of proIL-beta. Mol. Cell 2002, 10, 417–426.
- Kanneganti, T.D.; Ozoren, N.; Body-Malapel, M.; Amer, A.; Park, J.H.; Franchi, L.; Whitfield, J.; Barchet, W.; Colonna, M.; Vandenabeele, P.; et al. Bacterial RNA and small antiviral compounds activate caspase-1 through cryopyrin/Nalp3. Nature 2006, 440, 233–236.
- Mariathasan, S.; Weiss, D.S.; Newton, K.; McBride, J.; O’Rourke, K.; Roose-Girma, M.; Lee, W.P.; Weinrauch, Y.; Monack, D.M.; Dixit, V.M. Cryopyrin activates the inflammasome in response to toxins and ATP. Nature 2006, 440, 228–232.
- Martinon, F.; Petrilli, V.; Mayor, A.; Tardivel, A.; Tschopp, J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 2006, 440, 237–241.
- Franchi, L.; Amer, A.; Body-Malapel, M.; Kanneganti, T.D.; Ozoren, N.; Jagirdar, R.; Inohara, N.; Vandenabeele, P.; Bertin, J.; Coyle, A.; et al. Cytosolic flagellin requires Ipaf for activation of caspase-1 and interleukin 1beta in salmonella-infected macrophages. Nat. Immunol. 2006, 7, 576–582.
- Miao, E.A.; Alpuche-Aranda, C.M.; Dors, M.; Clark, A.E.; Bader, M.W.; Miller, S.I.; Aderem, A. Cytoplasmic flagellin activates caspase-1 and secretion of interleukin 1beta via Ipaf. Nat. Immunol. 2006, 7, 569–575.
- Mariathasan, S.; Newton, K.; Monack, D.M.; Vucic, D.; French, D.M.; Lee, W.P.; Roose-Girma, M.; Erickson, S.; Dixit, V.M. Differential activation of the inflammasome by caspase-1 adaptors ASC and Ipaf. Nature 2004, 430, 213–218.
- Xu, H.; Yang, J.; Gao, W.; Li, L.; Li, P.; Zhang, L.; Gong, Y.N.; Peng, X.; Xi, J.J.; Chen, S.; et al. Innate immune sensing of bacterial modifications of Rho GTPases by the Pyrin inflammasome. Nature 2014, 513, 237–241.
- Fernandes-Alnemri, T.; Yu, J.W.; Datta, P.; Wu, J.; Alnemri, E.S. AIM2 activates the inflammasome and cell death in response to cytoplasmic DNA. Nature 2009, 458, 509–513.
- Hornung, V.; Ablasser, A.; Charrel-Dennis, M.; Bauernfeind, F.; Horvath, G.; Caffrey, D.R.; Latz, E.; Fitzgerald, K.A. AIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature 2009, 458, 514–518.
- Cai, X.; Chen, J.; Xu, H.; Liu, S.; Jiang, Q.X.; Halfmann, R.; Chen, Z.J. Prion-like polymerization underlies signal transduction in antiviral immune defense and inflammasome activation. Cell 2014, 156, 1207–1222.
- Lu, A.; Magupalli, V.G.; Ruan, J.; Yin, Q.; Atianand, M.K.; Vos, M.R.; Schröder, G.F.; Fitzgerald, K.A.; Wu, H.; Egelman, E.H. Unified polymerization mechanism for the assembly of ASC-dependent inflammasomes. Cell 2014, 156, 1193–1206.
- Masumoto, J.; Taniguchi, S.; Ayukawa, K.; Sarvotham, H.; Kishino, T.; Niikawa, N.; Hidaka, E.; Katsuyama, T.; Higuchi, T.; Sagara, J. ASC, a novel 22-kDa protein, aggregates during apoptosis of human promyelocytic leukemia HL-60 cells. J. Biol. Chem. 1999, 274, 33835–33838.
- Sborgi, L.; Ravotti, F.; Dandey, V.P.; Dick, M.S.; Mazur, A.; Reckel, S.; Chami, M.; Scherer, S.; Huber, M.; Böckmann, A.; et al. Structure and assembly of the mouse ASC inflammasome by combined NMR spectroscopy and cryo-electron microscopy. Proc. Natl. Acad. Sci. USA 2015, 112, 13237–13242.
- Dinarello, C.A. Immunological and inflammatory functions of the interleukin-1 family. Annu. Rev. Immunol. 2009, 27, 519–550.
- Shi, J.; Zhao, Y.; Wang, K.; Shi, X.; Wang, Y.; Huang, H.; Zhuang, Y.; Cai, T.; Wang, F.; Shao, F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature 2015, 526, 660–665.
- He, W.T.; Wan, H.; Hu, L.; Chen, P.; Wang, X.; Huang, Z.; Yang, Z.H.; Zhong, C.Q.; Han, J. Gasdermin D is an executor of pyroptosis and required for interleukin-1beta secretion. Cell Res. 2015, 25, 1285–1298.
- Aglietti, R.A.; Estevez, A.; Gupta, A.; Ramirez, M.G.; Liu, P.S.; Kayagaki, N.; Ciferri, C.; Dixit, V.M.; Dueber, E.C. GsdmD p30 elicited by caspase-11 during pyroptosis forms pores in membranes. Proc. Natl. Acad. Sci. USA 2016, 113, 7858–7863.
- Ding, J.; Wang, K.; Liu, W.; She, Y.; Sun, Q.; Shi, J.; Sun, H.; Wang, D.C.; Shao, F. Pore-forming activity and structural autoinhibition of the gasdermin family. Nature 2016, 535, 111–116.
- Liu, X.; Zhang, Z.; Ruan, J.; Pan, Y.; Magupalli, V.G.; Wu, H.; Lieberman, J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature 2016, 535, 153–158.
- Sborgi, L.; Ruhl, S.; Mulvihill, E.; Pipercevic, J.; Heilig, R.; Stahlberg, H.; Farady, C.J.; Muller, D.J.; Broz, P.; Hiller, S. GSDMD membrane pore formation constitutes the mechanism of pyroptotic cell death. EMBO J. 2016, 35, 1766–1778.
- Kayagaki, N.; Stowe, I.B.; Lee, B.L.; O’Rourke, K.; Anderson, K.; Warming, S.; Cuellar, T.; Haley, B.; Roose-Girma, M.; Phung, Q.T.; et al. Caspase-11 cleaves gasdermin D for non-canonical inflammasome signalling. Nature 2015, 526, 666–671.
- Kayagaki, N.; Lee, B.L.; Stowe, I.B.; Kornfeld, O.S.; O’Rourke, K.; Mirrashidi, K.M.; Haley, B.; Watanabe, C.; Roose-Girma, M.; Modrusan, Z.; et al. IRF2 transcriptionally induces GSDMD expression for pyroptosis. Sci. Signal. 2019, 12, eaax4917.
- Benaoudia, S.; Martin, A.; Puig Gamez, M.; Gay, G.; Lagrange, B.; Cornut, M.; Krasnykov, K.; Claude, J.B.; Bourgeois, C.F.; Hughes, S.; et al. A genome-wide screen identifies IRF2 as a key regulator of caspase-4 in human cells. EMBO Rep. 2019, 20, e48235.
- Kerr, J.F.; Wyllie, A.H.; Currie, A.R. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257.
- Zou, H.; Henzel, W.J.; Liu, X.; Lutschg, A.; Wang, X. Apaf-1, a human protein homologous to C. elegans CED-4, participates in cytochrome c-dependent activation of caspase-3. Cell 1997, 90, 405–413.
- Kim, H.E.; Du, F.; Fang, M.; Wang, X. Formation of apoptosome is initiated by cytochrome c-induced dATP hydrolysis and subsequent nucleotide exchange on Apaf-1. Proc. Natl. Acad. Sci. USA 2005, 102, 17545–17550.
- Li, P.; Nijhawan, D.; Budihardjo, I.; Srinivasula, S.M.; Ahmad, M.; Alnemri, E.S.; Wang, X. Cytochrome c and dATP-dependent formation of Apaf-1/caspase-9 complex initiates an apoptotic protease cascade. Cell 1997, 91, 479–489.
- Boldin, M.P.; Goncharov, T.M.; Goltsev, Y.V.; Wallach, D. Involvement of MACH, a novel MORT1/FADD-interacting protease, in Fas/APO-1- and TNF receptor-induced cell death. Cell 1996, 85, 803–815.
- Muzio, M.; Chinnaiyan, A.M.; Kischkel, F.C.; O’Rourke, K.; Shevchenko, A.; Ni, J.; Scaffidi, C.; Bretz, J.D.; Zhang, M.; Gentz, R.; et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/APO-1) death--inducing signaling complex. Cell 1996, 85, 817–827.
- Gross, A.; Yin, X.M.; Wang, K.; Wei, M.C.; Jockel, J.; Milliman, C.; Erdjument-Bromage, H.; Tempst, P.; Korsmeyer, S.J. Caspase cleaved BID targets mitochondria and is required for cytochrome c release, while BCL-XL prevents this release but not tumor necrosis factor-R1/Fas death. J. Biol. Chem. 1999, 274, 1156–1163.
- Luo, X.; Budihardjo, I.; Zou, H.; Slaughter, C.; Wang, X. Bid, a Bcl2 interacting protein, mediates cytochrome c release from mitochondria in response to activation of cell surface death receptors. Cell 1998, 94, 481–490.
- Li, H.; Zhu, H.; Xu, C.J.; Yuan, J. Cleavage of BID by caspase 8 mediates the mitochondrial damage in the Fas pathway of apoptosis. Cell 1998, 94, 491–501.
- Wei, M.C.; Lindsten, T.; Mootha, V.K.; Weiler, S.; Gross, A.; Ashiya, M.; Thompson, C.B.; Korsmeyer, S.J. tBID, a membrane-targeted death ligand, oligomerizes BAK to release cytochrome c. Genes Dev. 2000, 14, 2060–2071.
- Kuwana, T.; Mackey, M.R.; Perkins, G.; Ellisman, M.H.; Latterich, M.; Schneiter, R.; Green, D.R.; Newmeyer, D.D. Bid, Bax, and lipids cooperate to form supramolecular openings in the outer mitochondrial membrane. Cell 2002, 111, 331–342.
- Gong, Y.; Fan, Z.; Luo, G.; Yang, C.; Huang, Q.; Fan, K.; Cheng, H.; Jin, K.; Ni, Q.; Yu, X.; et al. The role of necroptosis in cancer biology and therapy. Mol Cancer 2019, 18, 100.
- Nailwal, H.; Chan, F.K. Necroptosis in anti-viral inflammation. Cell Death Differ. 2019, 26, 4–13.
- Kang, T.B.; Ben-Moshe, T.; Varfolomeev, E.E.; Pewzner-Jung, Y.; Yogev, N.; Jurewicz, A.; Waisman, A.; Brenner, O.; Haffner, R.; Gustafsson, E.; et al. Caspase-8 serves both apoptotic and nonapoptotic roles. J. Immunol. 2004, 173, 2976–2984.
- Newton, K.; Wickliffe, K.E.; Dugger, D.L.; Maltzman, A.; Roose-Girma, M.; Dohse, M.; Kőműves, L.; Webster, J.D.; Dixit, V.M. Cleavage of RIPK1 by caspase-8 is crucial for limiting apoptosis and necroptosis. Nature 2019, 574, 428–431.
- O’Donnell, M.A.; Perez-Jimenez, E.; Oberst, A.; Ng, A.; Massoumi, R.; Xavier, R.; Green, D.R.; Ting, A.T. Caspase 8 inhibits programmed necrosis by processing CYLD. Nat. Cell Biol. 2011, 13, 1437–1442.
- Feng, S.; Yang, Y.; Mei, Y.; Ma, L.; Zhu, D.E.; Hoti, N.; Castanares, M.; Wu, M. Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cell. Signal. 2007, 19, 2056–2067.
- Grootjans, S.; Vanden Berghe, T.; Vandenabeele, P. Initiation and execution mechanisms of necroptosis: An overview. Cell Death Differ. 2017, 24, 1184–1195.
- Vandenabeele, P.; Galluzzi, L.; Vanden Berghe, T.; Kroemer, G. Molecular mechanisms of necroptosis: An ordered cellular explosion. Nat. Rev. Mol. Cell Biol. 2010, 11, 700–714.
- Sun, L.; Wang, H.; Wang, Z.; He, S.; Chen, S.; Liao, D.; Wang, L.; Yan, J.; Liu, W.; Lei, X.; et al. Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 2012, 148, 213–227.
- Zhao, J.; Jitkaew, S.; Cai, Z.; Choksi, S.; Li, Q.; Luo, J.; Liu, Z.G. Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc. Natl. Acad. Sci. USA 2012, 109, 5322–5327.
- Garcia, L.R.; Tenev, T.; Newman, R.; Haich, R.O.; Liccardi, G.; John, S.W.; Annibaldi, A.; Yu, L.; Pardo, M.; Young, S.N.; et al. Ubiquitylation of MLKL at lysine 219 positively regulates necroptosis-induced tissue injury and pathogen clearance. Nat. Commun. 2021, 12, 3364.
- Sharif, H.; Wang, L.; Wang, W.L.; Magupalli, V.G.; Andreeva, L.; Qiao, Q.; Hauenstein, A.V.; Wu, Z.; Núñez, G.; Mao, Y.; et al. Structural mechanism for NEK7-licensed activation of NLRP3 inflammasome. Nature 2019, 570, 338–343.
- Shi, H.; Wang, Y.; Li, X.; Zhan, X.; Tang, M.; Fina, M.; Su, L.; Pratt, D.; Bu, C.H.; Hildebrand, S.; et al. NLRP3 activation and mitosis are mutually exclusive events coordinated by NEK7, a new inflammasome component. Nat. Immunol. 2016, 17, 250–258.
- Schmid-Burgk, J.L.; Chauhan, D.; Schmidt, T.; Ebert, T.S.; Reinhardt, J.; Endl, E.; Hornung, V. A Genome-wide CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) Screen Identifies NEK7 as an Essential Component of NLRP3 Inflammasome Activation. J. Biol. Chem. 2016, 291, 103–109.
- He, Y.; Zeng, M.Y.; Yang, D.; Motro, B.; Nunez, G. NEK7 is an essential mediator of NLRP3 activation downstream of potassium efflux. Nature 2016, 530, 354–357.
- Samir, P.; Kesavardhana, S.; Patmore, D.M.; Gingras, S.; Malireddi, R.K.S.; Karki, R.; Guy, C.S.; Briard, B.; Place, D.E.; Bhattacharya, A.; et al. DDX3X acts as a live-or-die checkpoint in stressed cells by regulating NLRP3 inflammasome. Nature 2019, 573, 590–594.
- Kuriakose, T.; Zheng, M.; Neale, G.; Kanneganti, T.D. IRF1 Is a Transcriptional Regulator of ZBP1 Promoting NLRP3 Inflammasome Activation and Cell Death during Influenza Virus Infection. J. Immunol. 2018, 200, 1489–1495.
- Karki, R.; Lee, E.; Place, D.; Samir, P.; Mavuluri, J.; Sharma, B.R.; Balakrishnan, A.; Malireddi, R.K.S.; Geiger, R.; Zhu, Q.; et al. IRF8 Regulates Transcription of Naips for NLRC4 Inflammasome Activation. Cell 2018, 173, 920–933.e13.
- Kayagaki, N.; Kornfeld, O.S.; Lee, B.L.; Stowe, I.B.; O’Rourke, K.; Li, Q.; Sandoval, W.; Yan, D.; Kang, J.; Xu, M.; et al. NINJ1 mediates plasma membrane rupture during lytic cell death. Nature 2021, 591, 131–136.
- Christgen, S.; Place, D.E.; Kanneganti, T.D. Toward targeting inflammasomes: Insights into their regulation and activation. Cell Res. 2020, 30, 315–327.
- Bertheloot, D.; Latz, E.; Franklin, B.S. Necroptosis, pyroptosis and apoptosis: An intricate game of cell death. Cell Mol. Immunol. 2021, 18, 1106–1121.
- Holler, N.; Zaru, R.; Micheau, O.; Thome, M.; Attinger, A.; Valitutti, S.; Bodmer, J.L.; Schneider, P.; Seed, B.; Tschopp, J. Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat. Immunol. 2000, 1, 489–495.
- Oberst, A.; Dillon, C.P.; Weinlich, R.; McCormick, L.L.; Fitzgerald, P.; Pop, C.; Hakem, R.; Salvesen, G.S.; Green, D.R. Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 2011, 471, 363–367.
- Kaiser, W.J.; Upton, J.W.; Long, A.B.; Livingston-Rosanoff, D.; Daley-Bauer, L.P.; Hakem, R.; Caspary, T.; Mocarski, E.S. RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 2011, 471, 368–372.
- Alvarez-Diaz, S.; Dillon, C.P.; Lalaoui, N.; Tanzer, M.C.; Rodriguez, D.A.; Lin, A.; Lebois, M.; Hakem, R.; Josefsson, E.C.; O’Reilly, L.A.; et al. The Pseudokinase MLKL and the Kinase RIPK3 Have Distinct Roles in Autoimmune Disease Caused by Loss of Death-Receptor-Induced Apoptosis. Immunity 2016, 45, 513–526.
- Varfolomeev, E.E.; Schuchmann, M.; Luria, V.; Chiannilkulchai, N.; Beckmann, J.S.; Mett, I.L.; Rebrikov, D.; Brodianski, V.M.; Kemper, O.C.; Kollet, O.; et al. Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 1998, 9, 267–276.
- Lalaoui, N.; Boyden, S.E.; Oda, H.; Wood, G.M.; Stone, D.L.; Chau, D.; Liu, L.; Stoffels, M.; Kratina, T.; Lawlor, K.E.; et al. Mutations that prevent caspase cleavage of RIPK1 cause autoinflammatory disease. Nature 2020, 577, 103–108.
- Chun, H.J.; Zheng, L.; Ahmad, M.; Wang, J.; Speirs, C.K.; Siegel, R.M.; Dale, J.K.; Puck, J.; Davis, J.; Hall, C.G.; et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature 2002, 419, 395–399.
- Puri, A.W.; Broz, P.; Shen, A.; Monack, D.M.; Bogyo, M. Caspase-1 activity is required to bypass macrophage apoptosis upon Salmonella infection. Nat. Chem. Biol. 2012, 8, 745–747.
- Yu, J.; Nagasu, H.; Murakami, T.; Hoang, H.; Broderick, L.; Hoffman, H.M.; Horng, T. Inflammasome activation leads to Caspase-1-dependent mitochondrial damage and block of mitophagy. Proc. Natl. Acad. Sci. USA 2014, 111, 15514–15519.
- Pierini, R.; Juruj, C.; Perret, M.; Jones, C.L.; Mangeot, P.; Weiss, D.S.; Henry, T. AIM2/ASC triggers caspase-8-dependent apoptosis in Francisella-infected caspase-1-deficient macrophages. Cell Death Differ. 2012, 19, 1709–1721.
- Sagulenko, V.; Thygesen, S.J.; Sester, D.P.; Idris, A.; Cridland, J.A.; Vajjhala, P.R.; Roberts, T.L.; Schroder, K.; Vince, J.E.; Hill, J.M.; et al. AIM2 and NLRP3 inflammasomes activate both apoptotic and pyroptotic death pathways via ASC. Cell Death Differ. 2013, 20, 1149–1160.
- Man, S.M.; Tourlomousis, P.; Hopkins, L.; Monie, T.P.; Fitzgerald, K.A.; Bryant, C.E. Salmonella infection induces recruitment of Caspase-8 to the inflammasome to modulate IL-1β production. J. Immunol. 2013, 191, 5239–5246.
- Mascarenhas, D.P.A.; Cerqueira, D.M.; Pereira, M.S.F.; Castanheira, F.V.S.; Fernandes, T.D.; Manin, G.Z.; Cunha, L.D.; Zamboni, D.S. Inhibition of caspase-1 or gasdermin-D enable caspase-8 activation in the Naip5/NLRC4/ASC inflammasome. PLoS Pathog. 2017, 13, e1006502.
- Van Opdenbosch, N.; Van Gorp, H.; Verdonckt, M.; Saavedra, P.H.V.; de Vasconcelos, N.M.; Goncalves, A.; Vande Walle, L.; Demon, D.; Matusiak, M.; Van Hauwermeiren, F.; et al. Caspase-1 Engagement and TLR-Induced c-FLIP Expression Suppress ASC/Caspase-8-Dependent Apoptosis by Inflammasome Sensors NLRP1b and NLRC4. Cell Rep. 2017, 21, 3427–3444.
- Man, S.M.; Hopkins, L.J.; Nugent, E.; Cox, S.; Glück, I.M.; Tourlomousis, P.; Wright, J.A.; Cicuta, P.; Monie, T.P.; Bryant, C.E. Inflammasome activation causes dual recruitment of NLRC4 and NLRP3 to the same macromolecular complex. Proc. Natl. Acad. Sci. USA 2014, 111, 7403–7408.
- Kang, S.; Fernandes-Alnemri, T.; Rogers, C.; Mayes, L.; Wang, Y.; Dillon, C.; Roback, L.; Kaiser, W.; Oberst, A.; Sagara, J.; et al. Caspase-8 scaffolding function and MLKL regulate NLRP3 inflammasome activation downstream of TLR3. Nat. Commun. 2015, 6, 7515.
- Conos, S.A.; Chen, K.W.; De Nardo, D.; Hara, H.; Whitehead, L.; Nunez, G.; Masters, S.L.; Murphy, J.M.; Schroder, K.; Vaux, D.L.; et al. Active MLKL triggers the NLRP3 inflammasome in a cell-intrinsic manner. Proc. Natl. Acad. Sci. USA 2017, 114, E961–E969.
- Gutierrez, K.D.; Davis, M.A.; Daniels, B.P.; Olsen, T.M.; Ralli-Jain, P.; Tait, S.W.; Gale, M., Jr.; Oberst, A. MLKL Activation Triggers NLRP3-Mediated Processing and Release of IL-1β Independently of Gasdermin-D. J. Immunol. 2017, 198, 2156–2164.
- Sarhan, J.; Liu, B.C.; Muendlein, H.I.; Li, P.; Nilson, R.; Tang, A.Y.; Rongvaux, A.; Bunnell, S.C.; Shao, F.; Green, D.R.; et al. Caspase-8 induces cleavage of gasdermin D to elicit pyroptosis during Yersinia infection. Proc. Natl. Acad. Sci. USA 2018, 115, E10888–E10897.
- Demarco, B.; Grayczyk, J.P.; Bjanes, E.; Le Roy, D.; Tonnus, W.; Assenmacher, C.A.; Radaelli, E.; Fettrelet, T.; Mack, V.; Linkermann, A.; et al. Caspase-8-dependent gasdermin D cleavage promotes antimicrobial defense but confers susceptibility to TNF-induced lethality. Sci. Adv. 2020, 6, eabc3465.
- Orning, P.; Weng, D.; Starheim, K.; Ratner, D.; Best, Z.; Lee, B.; Brooks, A.; Xia, S.; Wu, H.; Kelliher, M.A.; et al. Pathogen blockade of TAK1 triggers caspase-8-dependent cleavage of gasdermin D and cell death. Science 2018, 362, 1064–1069.
- Taabazuing, C.Y.; Okondo, M.C.; Bachovchin, D.A. Pyroptosis and apoptosis pathways engage in bidirectional crosstalk in monocytes and macrophages. Cell Chem. Biol. 2017, 24, 507–514.
- Platnich, J.M.; Chung, H.; Lau, A.; Sandall, C.F.; Bondzi-Simpson, A.; Chen, H.M.; Komada, T.; Trotman-Grant, A.C.; Brandelli, J.R.; Chun, J.; et al. Shiga Toxin/Lipopolysaccharide Activates Caspase-4 and Gasdermin D to Trigger Mitochondrial Reactive Oxygen Species Upstream of the NLRP3 Inflammasome. Cell Rep. 2018, 25, 1525–1536.e1527.
- Rogers, C.; Erkes, D.A.; Nardone, A.; Aplin, A.E.; Fernandes-Alnemri, T.; Alnemri, E.S. Gasdermin pores permeabilize mitochondria to augment caspase-3 activation during apoptosis and inflammasome activation. Nat. Commun. 2019, 10, 1689.
- de Vasconcelos, N.M.; Van Opdenbosch, N.; Van Gorp, H.; Parthoens, E.; Lamkanfi, M. Single-cell analysis of pyroptosis dynamics reveals conserved GSDMD-mediated subcellular events that precede plasma membrane rupture. Cell Death Differ. 2019, 26, 146–161.
- Op de Beeck, K.; Van Camp, G.; Thys, S.; Cools, N.; Callebaut, I.; Vrijens, K.; Van Nassauw, L.; Van Tendeloo, V.F.; Timmermans, J.P.; Van Laer, L. The DFNA5 gene, responsible for hearing loss and involved in cancer, encodes a novel apoptosis-inducing protein. Eur. J. Hum. Genet. 2011, 19, 965–973.
- Wang, Y.; Gao, W.; Shi, X.; Ding, J.; Liu, W.; He, H.; Wang, K.; Shao, F. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature 2017, 547, 99–103.
- Rogers, C.; Fernandes-Alnemri, T.; Mayes, L.; Alnemri, D.; Cingolani, G.; Alnemri, E.S. Cleavage of DFNA5 by caspase-3 during apoptosis mediates progression to secondary necrotic/pyroptotic cell death. Nat. Commun. 2017, 8, 14128.
- Zhou, B.; Abbott, D.W. Gasdermin E permits interleukin-1 beta release in distinct sublytic and pyroptotic phases. Cell Rep. 2021, 35, 108998.
- Wang, C.; Yang, T.; Xiao, J.; Xu, C.; Alippe, Y.; Sun, K.; Kanneganti, T.D.; Monahan, J.B.; Abu-Amer, Y.; Lieberman, J.; et al. NLRP3 inflammasome activation triggers gasdermin D-independent inflammation. Sci. Immunol. 2021, 6, eabj3859.
- Tsuchiya, K.; Nakajima, S.; Hosojima, S.; Thi Nguyen, D.; Hattori, T.; Manh Le, T.; Hori, O.; Mahib, M.R.; Yamaguchi, Y.; Miura, M.; et al. Caspase-1 initiates apoptosis in the absence of gasdermin D. Nat. Commun. 2019, 10, 2091.
- Heilig, R.; Dilucca, M.; Boucher, D.; Chen, K.W.; Hancz, D.; Demarco, B.; Shkarina, K.; Broz, P. Caspase-1 cleaves Bid to release mitochondrial SMAC and drive secondary necrosis in the absence of GSDMD. Life Sci. Alliance 2020, 3, e202000735.
- Xu, W.; Che, Y.; Zhang, Q.; Huang, H.; Ding, C.; Wang, Y.; Wang, G.; Cao, L.; Hao, H. Apaf-1 Pyroptosome Senses Mitochondrial Permeability Transition. Cell Metab. 2021, 33, 424–436.e410.
- Chen, K.W.; Demarco, B.; Heilig, R.; Shkarina, K.; Boettcher, A.; Farady, C.J.; Pelczar, P.; Broz, P. Extrinsic and intrinsic apoptosis activate pannexin-1 to drive NLRP3 inflammasome assembly. EMBO J. 2019, 38, e101638.
- Yan, W.T.; Yang, Y.D.; Hu, W.M.; Ning, W.Y.; Liao, L.S.; Lu, S.; Zhao, W.J.; Zhang, Q.; Xiong, K. Do pyroptosis, apoptosis, and necroptosis (PANoptosis) exist in cerebral ischemia? Evidence from cell and rodent studies. Neural Regen. Res. 2022, 17, 1761–1768.
- Jiang, W.; Deng, Z.; Dai, X.; Zhao, W. PANoptosis: A New Insight Into Oral Infectious Diseases. Front. Immunol. 2021, 12, 789610.
- Chi, D.; Lin, X.; Meng, Q.; Tan, J.; Gong, Q.; Tong, Z. Real-Time Induction of Macrophage Apoptosis, Pyroptosis, and Necroptosis by Enterococcus faecalis OG1RF and Two Root Canal Isolated Strains. Front. Cell. Infect. Microbiol. 2021, 11, 720147.
- Lin, J.F.; Hu, P.S.; Wang, Y.Y.; Tan, Y.T.; Yu, K.; Liao, K.; Wu, Q.N.; Li, T.; Meng, Q.; Lin, J.Z.; et al. Phosphorylated NFS1 weakens oxaliplatin-based chemosensitivity of colorectal cancer by preventing PANoptosis. Signal Transduct. Target. Ther. 2022, 7, 54.
- Song, M.; Xia, W.; Tao, Z.; Zhu, B.; Zhang, W.; Liu, C.; Chen, S. Self-assembled polymeric nanocarrier-mediated co-delivery of metformin and doxorubicin for melanoma therapy. Drug Deliv. 2021, 28, 594–606.
- Upton, J.W.; Kaiser, W.J.; Mocarski, E.S. DAI/ZBP1/DLM-1 complexes with RIP3 to mediate virus-induced programmed necrosis that is targeted by murine cytomegalovirus vIRA. Cell Host Microbe 2012, 11, 290–297.
- Kaiser, W.J.; Upton, J.W.; Mocarski, E.S. Receptor-interacting protein homotypic interaction motif-dependent control of NF-kappa B activation via the DNA-dependent activator of IFN regulatory factors. J. Immunol. 2008, 181, 6427–6434.
- Thapa, R.J.; Ingram, J.P.; Ragan, K.B.; Nogusa, S.; Boyd, D.F.; Benitez, A.A.; Sridharan, H.; Kosoff, R.; Shubina, M.; Landsteiner, V.J.; et al. DAI senses influenza A virus genomic RNA and activates RIPK3-dependent cell death. Cell Host Microbe 2016, 20, 674–681.
- Wang, Y.; Kanneganti, T.D. From pyroptosis, apoptosis and necroptosis to PANoptosis: A mechanistic compendium of programmed cell death pathways. Comput. Struct. Biotechnol. J. 2021, 19, 4641–4657.
- Tamura, T.; Ishihara, M.; Lamphier, M.S.; Tanaka, N.; Oishi, I.; Aizawa, S.; Matsuyama, T.; Mak, T.W.; Taki, S.; Taniguchi, T. An IRF-1-dependent pathway of DNA damage-induced apoptosis in mitogen-activated T lymphocytes. Nature 1995, 376, 596–599.
- Tanaka, N.; Ishihara, M.; Kitagawa, M.; Harada, H.; Kimura, T.; Matsuyama, T.; Lamphier, M.S.; Aizawa, S.; Mak, T.W.; Taniguchi, T. Cellular commitment to oncogene-induced transformation or apoptosis is dependent on the transcription factor IRF-1. Cell 1994, 77, 829–839.
- Karki, R.; Kanneganti, T.D. The ‘cytokine storm’: Molecular mechanisms and therapeutic prospects. Trends Immunol. 2021, 42, 681–705.
- Man, S.M.; Karki, R.; Malireddi, R.K.; Neale, G.; Vogel, P.; Yamamoto, M.; Lamkanfi, M.; Kanneganti, T.D. The transcription factor IRF1 and guanylate-binding proteins target activation of the AIM2 inflammasome by Francisella infection. Nat. Immunol. 2015, 16, 467–475.
