Human milk (HM) is considered the gold standard for infant nutrition. HM contains macro- and micronutrients, as well as a range of bioactive compounds (hormones, growth factors, cell debris, etc.). The analysis of the complex and dynamic composition of HM has been a permanent challenge for researchers. The use of novel, cutting-edge techniques involving different metabolomics platforms has permitted to expand knowledge on the variable composition of HM. Here, the state-of-the-art in untargeted metabolomic studies of HM, with emphasis on sampling, extraction and analysis steps is presented.
- human milk
- metabolome
- sampling
- extraction
- liquid chromatography–mass spectrometry
1. Introduction
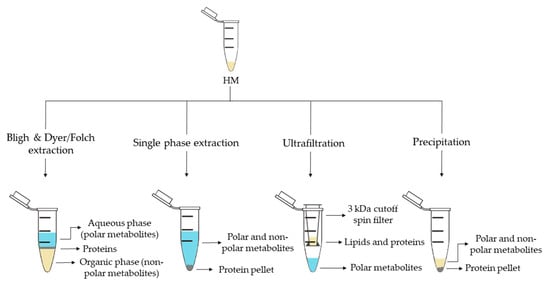
| Sample Preparation (1st. step) | Sample Preparation (2nd. step) | Compound Class | Platform | Column/Capillary | References |
|---|---|---|---|---|---|
| Bligh & Dyer extraction | Deuterated solvent addition to aqueous phase | Polar metabolites | 1H-NMR | - | [13]][13[16],16[29][32,29,32] |
| Derivatization of aqueous phase: methoximation and silylation | Polar metabolites and FAs | GC-MS | DB-5ms | [17][18][19][17,18,19] | |
| Derivatization of organic phase: methylation | FAs | GC-MS | DB-5ms | [13] | |
| Direct injection of aqueous phase | Polar metabolites | LC-QTOF-MS (+) | HILIC | [35] | |
| Redissolution of aqueous phase in H2O:ACN (95:5) | Polar metabolites | LC-Orbitrap-MS (+, −) | C18 | [24] | |
| Redissolution of organic phase in (ACN:IPA:H2O (65:30:5) |
Lipidic metabolites | LC-Orbitrap-MS (+,−) | C18 | [25] | |
| Folch extraction | Deuterated solvent addition to aqueous and organic phases | Hydrophobic and polar metabolites | 1H-NMR | - | [28] |
| Redissolution of aqueous phase in formic acid and centrifugation | Polar metabolites (amino acids) | CE-TOF-MS (+) | 60 m × 50 µm I.D. | ||
| Redissolution of organic phase in IPA:H2O:ACN (2:1:1) and centrifugation | Lipidic metabolites | UPLC-QTOF-MS (+,−) | C18 | ||
| Single phase extraction | Derivatization: methoximation and silylation | Polar metabolites and FAs | GC-MS | DB-5ms | [27][28][27,28] |
| Direct injection | Lipidic (and polar) metabolites | LC-QTOF-MS (+,−) | C8 | [27][28][27,28] | |
| UPLC-QTOF-MS (+) | C18 | [15] | |||
| Fat extraction with n-hexane/IPA | Deuterated solvent addition | TGs | 13C-NMR; 1H-NMR | - | [20] |
| Filtration 3 kDa cutoff spin filter | Deuterated solvent addition | Polar metabolites | 1H-NMR | - | [14][21][22][29][33][14,21,22,29,33] |
| Protein precipitation | Derivatization: methoximation and silylation | Polar metabolites | GC-MS | DB-5ms | [36] |
| Hybrid SPE-Phospholipid extraction and redissolution in diluted organic phase of Bligh & Dyer extraction | Lipidic metabolites | LC-QTOF-MS (+) | C8 | [35] | |
| Fat removal with CH2Cl2 and dansylation of aqueous phase | Polar metabolites (amine/phenol submetabolome) | Chemical isotope labelling LC-QTOF-MS (+) | C18 | [42][43][45,46] | |
| Direct injection | Polar metabolites and FAs | UPLC-QTOF-MS (+,−) | C18 | [18] | |
| Fat removal by centrifugation | Two additional centrifugations and deuterated solvent addition | Polar metabolites | 1H-NMR | - | [34] |
| Filtration 10 kDa cutoff spin filter and deuterated solvent addition | Polar metabolites | 1H-NMR | - | [23][26][23,26] | |
| Homogenization | Deuterated solvent addition | Polar metabolites | 1H-NMR | - | [31] |
| H2O-dilution | NaBH4-reduction and PGC cartridge | Oligosaccharides | UPLC-TQD-MS (+) | Hypercarb® | [24] |
3. The HM Metabolome: Compound Annotation and Coverage
As in other areas of metabolomic research, compound identification is still a major bottleneck in data analysis and interpretation. The Metabolomics Standards Initiative’s (MSI) defines four levels of metabolite identification, which include: identified metabolites (level 1); putatively annotated compounds (level 2); putatively annotated compound classes (level 3); and unknown compounds (level 4) [44][60]. Due to the limited availability of pure analytical standards required to reach level 1, biological databanks and spectral databases are the most important resources for metabolite annotation (levels 2 and 3). A large number of databases are available today, providing different levels of information and complementary data on chemical structures, physicochemical properties, biological functions, and pathway mapping of metabolites [45][61]. The metabolomics community classifies these resources in several categories: (i) chemical databases; (ii) spectral libraries; (iii) pathway databases; (iv) knowledge databases; and (v) references repositories [46][62]. Regarding HM metabolomics, the most frequently used databases and libraries are: Human Metabolome Database (HMDB) [47][63], Metabolite and Chemical Entity Database (METLIN) [48][64], National Institute of Science and Technology (NIST) library, Fiehn RTL Library [49][65], LipidMAPS Structure Database (LMSD) [50][66], Milk Metabolome Database (MCDB) [51][52][67,68], Kyoto Encyclopedia of Genes and Genomes (KEGG) [53][69], MycompoundID with the evidence-based metabolome library (EML) [54][70], Chenomx NMR Suite Profiles and other online university databases, such as CEU-mass mediator [55][56][71,72]. Metabolite assignment in NMR spectra has been performed based on literature data and commercial resonance databases, such as Chenomx NMR Suite Profiles. Metabolite annotation was contrasted with in-house libraries containing pure compound spectra. Some of the proposed assignments were confirmed by two-dimensional NMR spectra, such as Correlation Spectroscopy (COSY) [13][29][31][32][13,29,31,32], Homonuclear Correlation Spectroscopy (TOCSY) [13][31][32][34][13,31,32,34], Diffusion-Ordered Spectroscopy (DOSY) [32], Heteronuclear Single Quantum Coherence Spectroscopy (HSQC) [32][34][32,34], and Heteronuclear Multiple Bond Correlation (HMBC) [32]. In LC-MS and CE-MS-based studies of the HM metabolome, tentative metabolite annotation has been carried out by matching of accurate masses, isotopic profiles, and/or fragmentation patterns to candidate metabolites in online databases such as KEGG, METLIN, LipidMAPS, and HMDB [18][24][25][27][28][35][18,24,25,27,28,35]. In-house built databases generated by the analysis of commercial standards are also commonly employed [24][25][24,25]. In GC-MS, retention index (RI) corrections are made by analyzing a fatty acid methyl ester (FAME) mixture standard solution and assigning a match score between the experimental FAME mixture and theoretical RI values based on the values contained in the Fiehn RTL library. Furthermore, metabolites were complementarily annotated by comparing their mass fragmentation patterns with those available in Fiehn RTL and NIST libraries [13][17][18][19][27][28][36][13,17,18,19,27,28,36]. A comprehensive list of annotated and/or identified metabolites in HM from untargeted metabolomics studies [14][15][17][18][19][21][22][23][24][25][26][27][28][29][31][32][33][34][35][36][14,15,17,18,19,21,22,23,24,25,26,27,28,29,31,32,33,34,35,36] is reported in Table S1. This table contains information about the metabolites reported in each reference, such as their molecular formula, IDs (LipidMAPS and/or HMDB IDs), the extraction procedure performed, the analytical platform used, and the detected metabolite class. Readers can select metabolites dynamically by filtering data according to the latter information. A total of 1187, 111, and 128 metabolites were reported using LC-MS, GC-MS, and NMR, respectively (see Figure 24). As shown in the Venn diagram, LC-MS and GC-MS allowed the detection of 36 common metabolites (mainly carbohydrates and FAs); a total of 29 metabolites overlapped between LC-MS and NMR (principally oligosaccharides); and 21 metabolites (predominantly amino acids and organic acids) were commonly reported in GC-MS and NMR based studies. Only 13 metabolites were reported by all three platforms, i.e., creatine, tyrosine, arabinose, galactose, glucose, lactose, maltose, capric acid/caprate, caprylic acid/ caprylate, citric acid/citrate, pyruvic acid/pyruvate, hippuric acid/hippurate, and myo-inositol. These metabolites were assigned to different classes including amino acids, carbohydrates, FAs, and organic acids.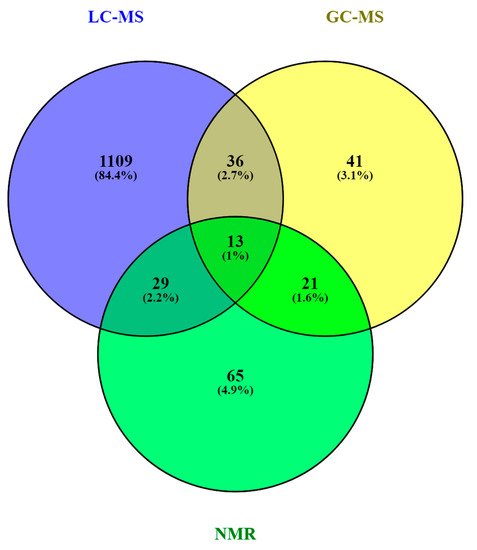
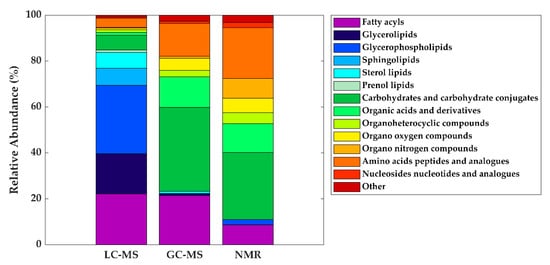
| Metabolite class | LC-MS | GC-MS | NMR |
|---|---|---|---|
| Fatty acyls | Linoleic acid (C18:2) | Oleic acid (C18:1) | - |
| Oleic acid (C18:1) | Palmitic acid (C16:0) | ||
| Palmitoleic acid (C16:1) | Stearic acid (C18:0) | ||
| Glycerolipids | DG (36:1) | - | - |
| Glycerophospholipids | LysoPC (16:0) | - | - |
| Carbohydrates and carbohydrate conjugates | - | Fructose | Lactose |
| Fucose | |||
| Ribose | |||
| Organic acids and derivatives | - | Malic acid Urea |
Acetate |
| Citrate | |||
| Lactate | |||
| Organo nitrogen compounds | - | - | Choline |
| Amino acids, peptides, and analogues | - | Alanine Glutamate Glycine Pyroglutamic acid Serine Valine |
Alanine Creatine Glutamate Glutamine Isoleucine Leucine Tyrosine Valine |
4. Conclusions and Future Perspectives
In less than a decade, 26 research papers have been published trying to shed light on the complex and dynamic composition of HM and the feasibility of different options for sample extraction and metabolite detection has been demonstrated. Due to the many factors that influence HM composition, a thorough study design including SOPs for milk extraction, collection, and storage is indispensable for obtaining biologically meaningful results. Multi-platform approaches are encouraged for providing adequate metabolome coverage, as the diversity of compounds contained in HM will not be properly reflected using one single assay. In line with metabolomics workflows tailored to other sample types, the reproducibility of HM metabolomics studies will benefit from the implementation of QA/QC procedures. Automated metabolite annotation and identification with pure chemical standards is warranted and the authors encourage the use of publicly accessible platforms for enabling the exchange of raw data for comparison between studies.
