Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Yeh-Han Wang and Version 3 by Camila Xu.
Tumor mutational burden (TMB) refers to the total load of somatic mutations in tumor cells. It has been approved as a predictive biomarker for immune checkpoint inhibitors (ICIs), next-generation sequencing (NGS) TMB panels are being increasingly used clinically.
- tumor mutational burden (TMB)
- next-generation sequencing (NGS)
- harmonization
1. Introduction
Along with the investigation of immune checkpoint inhibitors (ICIs), tumor mutational burden (TMB) has been developed to be a predictive biomarker in recent years. By definition, TMB refers to the total load of somatic mutations in tumor cells. As somatic mutations may cause specific tumor neoantigens, patients with a high TMB are likely to be responsive to immunotherapy [1][2][3][4][1,2,3,4]. A high TMB was first noted to be associated with the treatment response of cytotoxic T-lymphocyte-associated protein 4 (CTLA-4) inhibitors in melanoma [5][6][5,6]. In the following years, TMB was employed in many clinical trials of anti-programmed cell death protein 1 (PD-1)/programmed cell death protein-ligand 1 (PD-L1) agents to treat various cancer types. Patients with a higher TMB tended to exhibit a better treatment response, but the testing methods and cutoffs of TMB varied across trials [3][7][8][9][10][11][12][13][14][3,7,8,9,10,11,12,13,14].
Several sequencing methods and multi-gene panels have been established to test TMB in academic, medical and diagnostic laboratories. Originally, the gold standard to calculate TMB was whole-exome sequencing (WES), where the total number of somatic mutations was calculated and reported. However, this is less feasible in most clinical settings because of its labor intensiveness and high cost, the lengthy turnaround time and the lack of computational or bioinformatics specialists. Panel-based TMB assays were thus developed by many laboratories and diagnostic companies. Generally, a TMB panel includes several hundreds of genes, and the somatic mutation load in tumors is estimated using specified bioinformatics algorithms. However, as the design varies across panels from gene selection to bioinformatics algorithms, no universal cutoff defines a high TMB status. The variation between TMB estimates can confuse clinicians and may hinder clinical decision making. Further harmonization and standardization are mandatory for the clinical implementation of panel-based TMB assays.
The standard is still unclear and sometimes confusing, although several TMB panels have been approved or cleared by the U.S. Food and Drug Administration (FDA). In the KEYNOTE-158 study, TMB was defined as a predictive biomarker, and a cutoff of 10 mutations per megabase (muts/Mb) to define a high TMB was proposed and further approved by the FDA as a tumor agnostic indication for the prescription of pembrolizumab [9][15][9,15]. A commercial laboratory-based panel, FoundationOne (F1) CDx (Foundation Medicine Inc.), was simultaneously approved as the companion diagnostic test for this indication. In addition, the Memorial Sloan Kettering Cancer Center (MSKCC) also developed an in-house cancer genomic profiling assay, MSK-IMPACT (Integrated Mutation Profiling of Actionable Cancer Targets), which was cleared through the FDA 510(k) review in 2017 [16][17][16,17]. In the following years, several other next-generation sequencing (NGS) panels also obtained approval from the FDA. However, it is difficult to harmonize the inter-panel variation between NGS panels, regardless of whether they have regulatory authorization or clinical validation data from trials.
2. Harmonization and Standardization of Panel-Based TMB
2.1. Preanalytic Factors (Sample and DNA Issues)
Several preanalytic factors may influence the TMB estimation, primarily associated with DNA quality. Poor DNA quality, whether resulting from specimen handling, processing or archiving, is known to cause more false-positive calls of somatic mutations, which often feature a low allele frequency [18][30]. These false-positive mutations inappropriately increase the TMB estimates. Quy et al. noted that patients with high TMB estimates from specimens with a low DNA library concentration show no to less treatment benefits from anti-PD-L1 antibodies compared to the high concentration group [19][23]. While a low library concentration reflects poor DNA quality, the high TMB status determined by testing such specimens was more likely to be misclassified. In this researchtudy, significant increases in false-positive variant calls were also observed in formalin-fixed paraffin-embedded (FFPE) tissue specimens compared to frozen-fixed fresh tissue, a phenomenon that suggests that formalin potentially damages DNA tissue, especially with improper fixation [20][22]. Formalin-fixation-induced deamination is one of the preanalytic factors that leads to the overestimation of TMB. Tumor fraction and input DNA quantity both affect the sensitivity of TMB testing, though their impacts vary in degrees between cancer types and panels. Some cancer types are known to have a higher TMB, such as NSCLC and melanoma, while others mostly express a low TMB. Generally, a low tumor fraction may cause missed calls of tumor somatic mutation and, thus, underestimation of TMB. However, panel-specific bioinformatics algorithms may partially compensate for these effects, and an acceptance criterion of tumor fraction has been verified in some TMB panels. The effects of a low tumor fraction and DNA input can be minimized through routine measures of laboratory quality assurance/quality control (QA/QC).2.2. Sequencing Factors
Gene coverage and panel size are key factors affecting the performance of a TMB panel. Genes selected in TMB panels, together with their bioinformatics algorithm, primarily determine the accuracy and variability in TMB estimates [21][40]. As the genes selected in a panel are mostly cancer-related, their distribution and coverage in the genome are not randomly or evenly distributed. In addition, the prevalence of cancer gene mutations is also different across cancer types. Therefore, gene selection may cause internal bias for panel-based TMB estimates, which requires further calibration by the bioinformatics algorithm. Generally, the combination of gene selection and bioinformatics has been optimized in most laboratories and assays, and it has demonstrated comparability with WES TMB. However, the phase II FoCR harmonization study observed a tendency toward the overestimation of TMB when known pathogenic cancer genes were not excluded from the estimation [22][20]. Panel size was also critical to determine the variability in a TMB assay. Several studies indicate that smaller panels (<1 Mb) exhibit significant variability when correlating with WES TMB calculation [23][24][25][26][24,41,42,43].2.3. Bioinformatics Factors
The initial clinical investigation of TMB was to calculate the sum of somatic mutations detected by WES. Several calculation strategies based on the inclusion types of mutations have been developed in different studies. Although attempts to include all somatic mutation types (e.g., synonymous, non-synonymous single-nucleotide variations (SNVs) and small insertions and deletions (indel)) for TMB calculations have been reported in several studies [14][27][28][29][30][14,45,46,47,48], the calculation that includes missense (non-synonymous) mutations only has been the mainstay approach for WES TMB [31][26]. Chang et al. investigated the impacts of the two calculation strategies above on the TMB value using CheckMate 026 data and found a perfect correlation between “all mutation types” and “missense mutation only” (Spearman’s rho = 0.99), but the former calculation exhibited 3.1-fold higher TMB values compared to the latter (median TMB: 540 muts for all mutation types while 170 muts for missense mutation only) [31][26]. As TMB panels only detect hundreds of genes (approximately accounting for 0.5–2 megabase of the genome across panels), various computational approaches have been optimized across laboratories or commercialized panels for a better correlation between panel-based TMB and WES TMB [26][43]. The core steps of the panel-based TMB calculation are shown in Figure 1.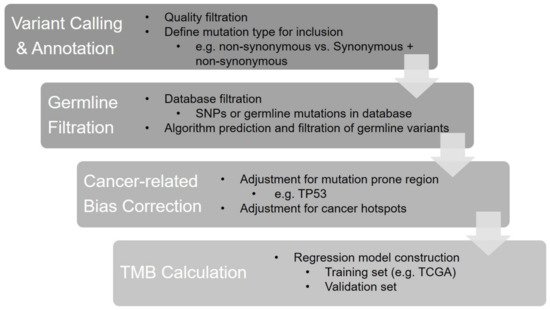
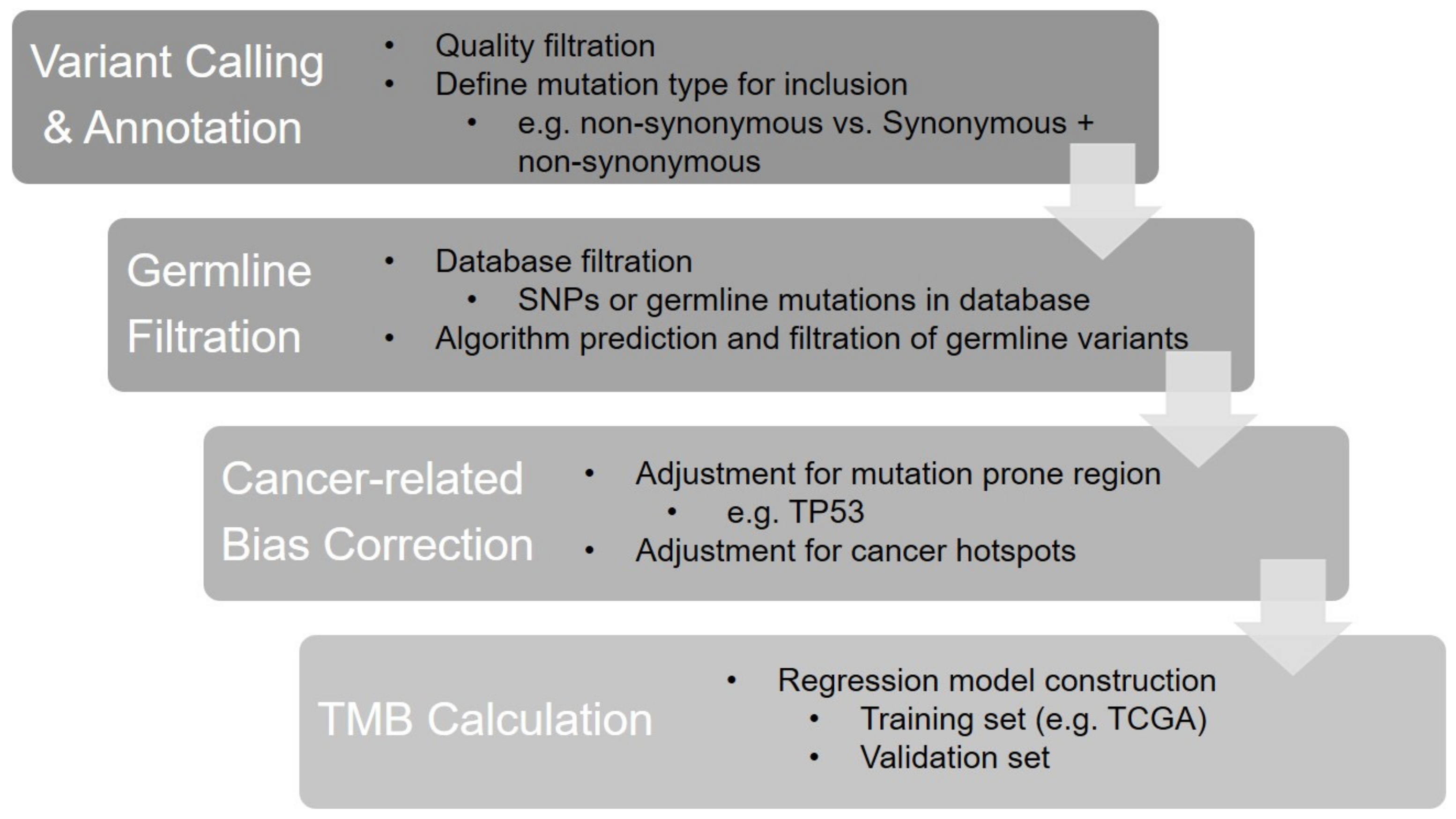
Figure 1. The bioinformatics algorithm of panel-based TMB calculation.
