Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Catherine Yang and Version 1 by Yanzhen Bi.
In mammalian cells, double-strand breaks (DSBs) are repaired predominantly by error-prone non-homologous end joining (NHEJ), but less prevalently by error-free template-dependent homologous recombination (HR). DSB repair pathway selection is the bedrock for genome editing. NHEJ results in random mutations when repairing DSB, while HR induces high-fidelity sequence-specific variations, but with an undesirable low efficiency.
- DSB repair
- NHEJ
- Genome editing
- RNA template
1. DSB Induction and Pathway Choice
In mammalians, genome editing through CRISPR (clustered regularly interspaced short palindromic repeats)-Cas (CRISPR-associated) nuclease systems can induce double-strand breaks (DSBs), initiating one of the conserved repertoire repair pathways, depending on the type of damage, cellular context, and phase of the cell cycle [1,2][1][2]. CRISPR/Cas9 genome editing actually takes advantage of DSB repair pathways to introduce variations into the user-defined genomic loci. Eukaryotes possess highly coordinated repair processes, and DSBs can be repaired through non-homologous end joining (NHEJ) or homologous recombination (HR) pathways. Broken DSB ends activate phophotidylinositol-3 kinase-like kinases (PIKKs) (Figure 1a), which phosphorylates histone H2AX (known as γH2AX) leading to cell cycle arrest, a pause in the local transcription of RNA polymerase-I (RNAP-I), RNA polymerase-II (RNAP-II) activation, and up-regulation of genes encoding DNA damage repair factors [3,4,5,6][3][4][5][6]. γH2AX facilitates in the activation of NHEJ through 53BP1 and KU (Ku70/Ku80) heterodimers [7,8[7][8][9],9], or it can interact with NBS1, through the localization of the MRN (MRE11-RAD50-NBS1) and BRCA1-CtIP (C-terminal binding protein interacting protein) complex at the DSB site to initiate HR repair pathway, as shown in Figure 1b [1].
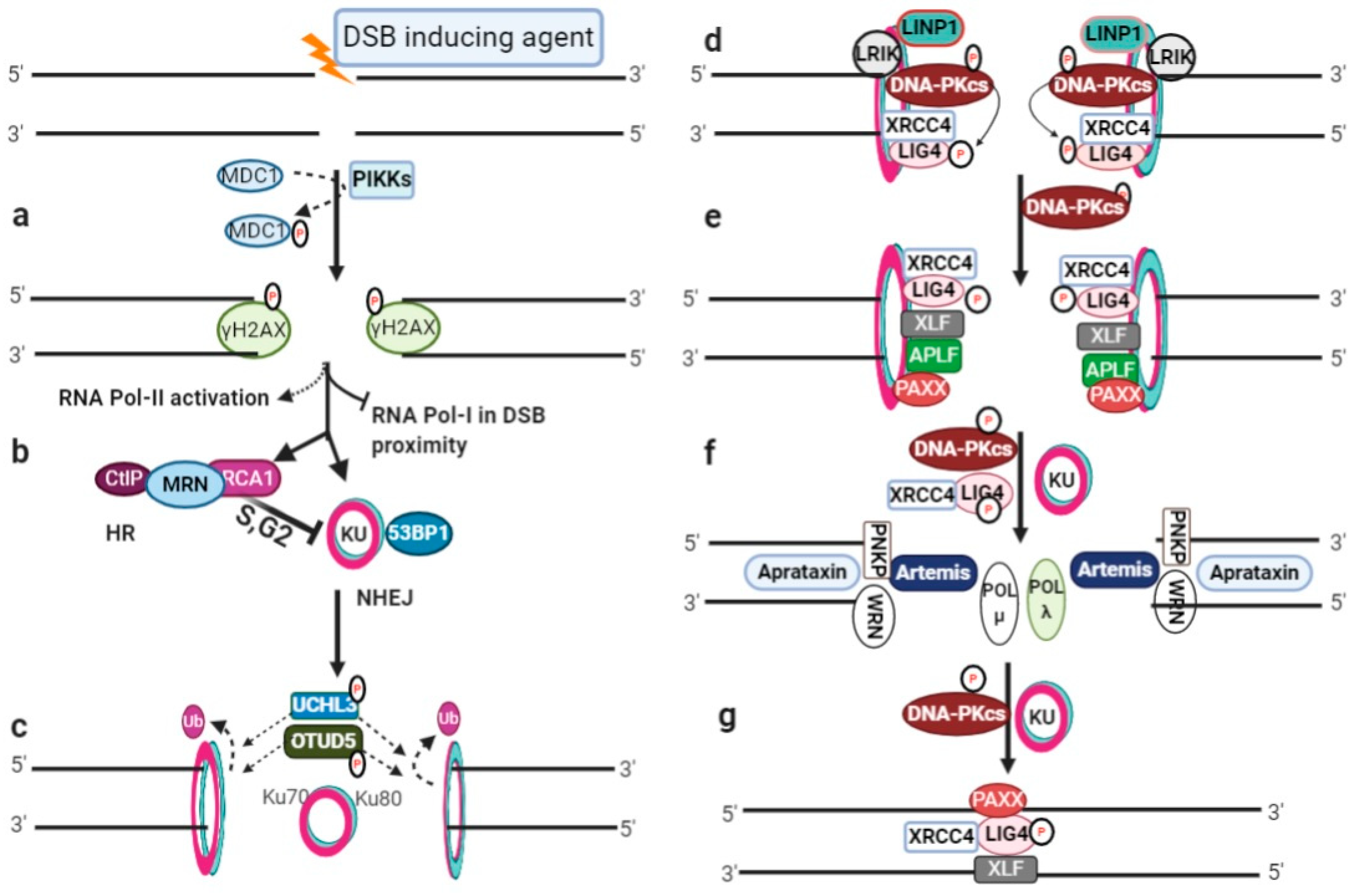
Figure 1. DSB induction and NHEJ repair pathway: (a) DSB initiates phosphorylation by activated phophotidylinositol-3 kinase-like kinases (PIKKs) and phosphorylation of H2AX (γH2AX) for cascade signaling; (b) RNA polymerase I-based local transcription inhibition in the DSB proximity, RNA polymerase II activation for damage-induced RNA generation, and the recruitment of NHEJ or HR repair machinery; (c) KU heterodimer binding at broken ends for NHEJ repair initiation and deubiquitylation of the Ku80 subunit; (d) DNA-PKcs autophosphorylates and then phosphorylates LIG4. LINP1 and LRIK are non-coding RNAs that stabilize the KU–DNA complex; (e) recruitment of XLF, APLF, and PAXX; (f) end processing by Artemis nuclease and other end processing molecules, and gap filling by polymerases; (g) LIG4 ligate-processed ends assisted with XRCC4, PAXX and XLF.
2. Non-Homologous End Joining (NHEJ)
NHEJ is the predominant, template-independent repair pathway and can potentially re-ligate any type of damaged DNA ends throughout the cell cycle [2,3][2][3]. NHEJ can be subdivided into three main sub-sequential steps, namely, (i) DSB end recognition and recruitment of NHEJ machinery, (ii) DNA ends processing, and (iii) DNA ends ligation. The schematic presentation of NHEJ repair pathway is illustrated in Figure 1c–g.
3. Homologous Recombination (HR)
In eukaryotes, accurate DSB repair is performed by HR, which requires sister-chromatid/repair-template generally restricted to the S/G2 phase of the cell cycle. HR can be categorized into subsections (i) DNA ends resection, (ii) RAD51-nucleofilament formation, (iii) homology searching, (iv) hDNA formation and D-loop extension, and (vi) D-loop dissolution.4. Roles of RNA Transcripts in DSB Repair
4.1. dincRNAs in DSB Repair
DSB induction inhibits RNA polymerase I (RNAP-I) to down-regulate ongoing local transcription, while it activates RNAP-II promoter-independent activity to generate damage-induced ncRNAs (dincRNA), as shown in Figure 1 and Figure 32 [141][10]. These dincRNAs can be categorized on the basis of size into long (dilncRNA) >200 nt and small ncRNAs with about 21 nt [142][11]. Long ncRNAs (lncRNAs) act as a precursor for small ncRNAs, analogous to miRNA biosynthesis [143][12]. These dincRNAs are exported to the cytoplasm via RNA binding proteins (RBPs), and protect the cell by degrading truncated mRNA [142,144,145][11][13][14]. The role of dincRNAs in the DSB repair process is controversial, as Michelini et al. identified that the MRN complex recruits RNAP-II in the DSB vicinity for DICER or AGO2-mediated HR repair [142][11]. However, DICER inactivation is reported to reduce 53BP1 foci formation, suggesting its role in the NHEJ repair pathway [146][15]. Similarly, pre-existing dincRNA transcripts have been reported prior to DSB induction by Bader and Bushell [147][16], indicating that additional studies are needed in order to understand the detailed molecular mechanisms.
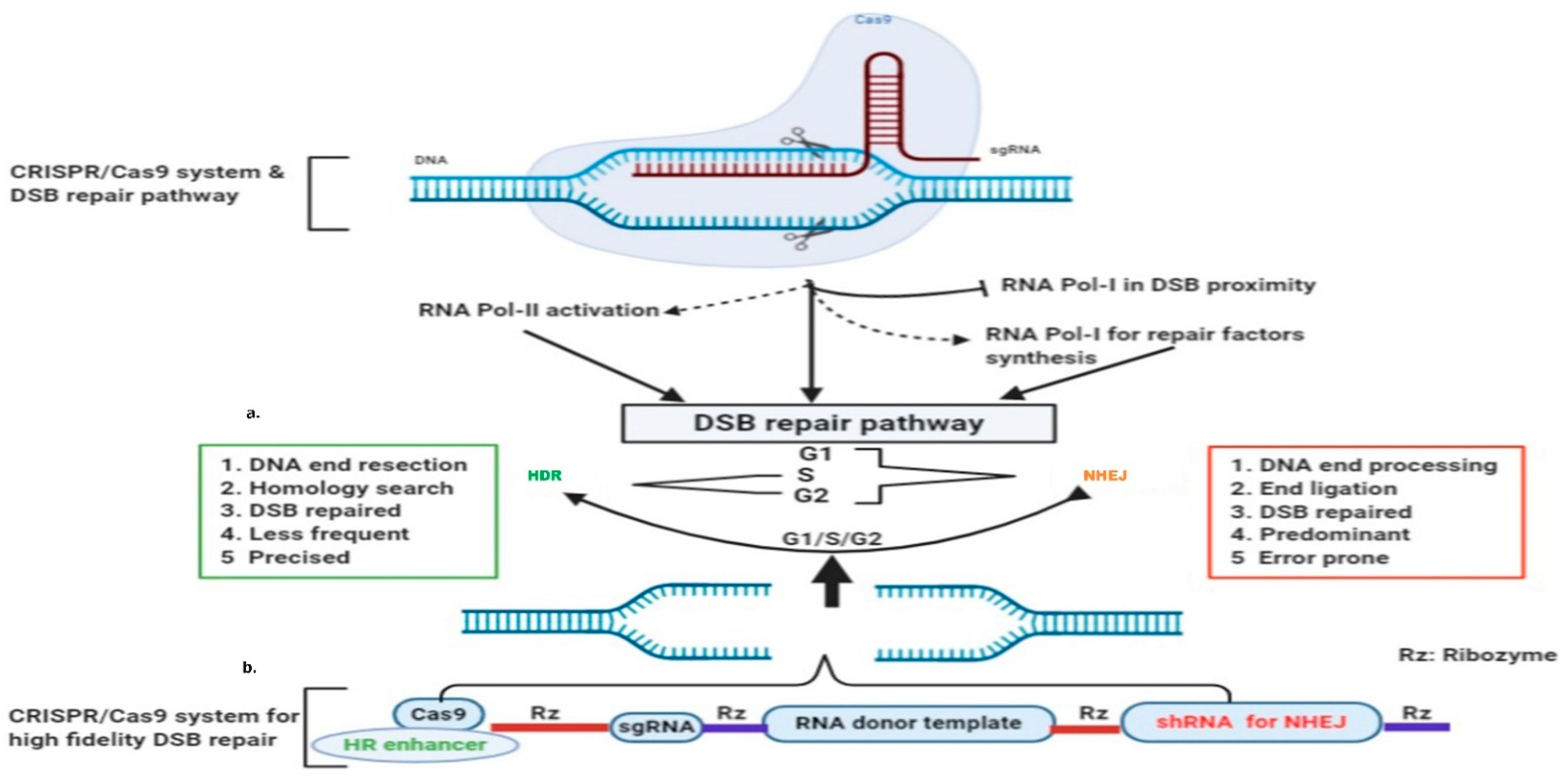
Figure 32. CRISPR-Cas9 induced DSB and RNA-templated high-fidelity repair system: (a) Cas9 induced broken ends pause RNA polymerase-I transcription in the DSB vicinity, and activate RNA Polymerase-II and RNA polymerase-I for the synthesis of DSB repair factors; (b) single construct for Cas9 expression ribozyme mediated transcription of sgRNA, donor template with desired sequence, and shRNA for NHEJ suppression.
4.2. RNA-Template Mediated DSB Repair
The RNAP-I transcription pause transcribing DSB region is essential to halt truncated mRNA synthesis. Moreover, it is also important, as nascent RNA is proposed to serve as a template [148][17]. As proposed by Lavigne et al., nascent transcripts or pre-mRNAs are available for transcript-templated repair shortly after the initiating lesion [149][18]. It is widely accepted that RNAs can pair with homologous DNA strands to form RNA–DNA hybrids [144,150][13][19]. RNA-binding proteins (RBPs) such as THRAP3, DICER, DHX9, Drosha, and Senataxin are important driving factors [146,151,152,153,154][15][20][21][22][23]. These possess different functions, for example Drosha promotes RNA–DNA hybrids, while Senataxin and RNase H2 resolve [144,154,155,156][13][23][24][25]. A plethora of studies indicate that RNA–DNA hybrids have direct roles in DSB repair [145,157,158][14][26][27]. Mazina et al. revealed that RPA has a higher RNA binding affinity than DNA, and RAD52 is able to invert the strand exchange between homologous DNA and RNA [159][28]. It is evident that defective H-RNases could stimulate homologous RNA-mediated DNA repair more than the homologous DNA-template in the presence of RPA [160][29]. The emerging picture is that dincRNAs are important for truncated mRNA degradation as well as cascade signaling, and RNA molecules homologous to DSB ends could serve as a template for faithful repair through HR. An interesting finding about DNA polymerase θ (Polθ) has revealed its reverse transcription ability, similar to reverse transcriptases in retrovirus. Polθ is efficient in the incorporation of deoxyribonucleotides on RNA versus DNA, and promotes RNA-templated DNA repair in mammalian cells [161][30]. This suggests the significance of RNA as a template in DNA repair.
4.3. RNA versus DNA as a Donor Repair Template
Both single-stranded oligonucleotides (ssODNs) and double-stranded DNA (dsDNA) are used in genome editing (GE). ssODNs provide higher insertion efficiencies over the dsDNA template [162][31]. However, ssODNs require efficient delivery in the nuclear region to avoid degradation from ssDNAases [163][32]. ssODNs can also generate null alleles and are inefficient when applied at scale [164][33]. The other main disadvantage of ssODNs is their size limitation. Although limited data are available, it has been demonstrated that DSBs can be repaired via the RNA donor template repair pathway [160,165,166][29][34][35].
References
- Kobayashi, J.; Tauchi, H.; Sakamoto, S.; Nakamura, A.; Morishima, K.; Matsuura, S.; Kobayashi, T.; Tamai, K.; Tanimoto, K.; Komatsu, K. NBS1 localizes to gamma-H2AX foci through interaction with the FHA/BRCT domain. Curr. Biol. 2002, 12, 1846–1851.
- Davis, A.J.; Chen, D.J. DNA double strand break repair via non-homologous end-joining. Transl. Cancer Res. 2013, 2, 130–143.
- Reid, D.A.; Keegan, S.; Leo-Macias, A.; Watanabe, G.; Strande, N.T.; Chang, H.H.; Oksuz, B.A.; Fenyo, D.; Lieber, M.R.; Ramsden, D.A.; et al. Organization and dynamics of the nonhomologous end-joining machinery during DNA double-strand break repair. Proc. Natl. Acad. Sci. USA 2015, 112, E2575–E2584.
- Downs, J.A.; Jackson, S.P. A means to a DNA end: The many roles of Ku. Nat. Rev. Mol. Cell Biol. 2004, 5, 367–378.
- Fell, V.L.; Schild-Poulter, C. The Ku heterodimer: Function in DNA repair and beyond. Mutat. Res. Rev. Mutat. Res. 2015, 763, 15–29.
- Zhang, Z.; Hu, W.; Cano, L.; Lee, T.D.; Chen, D.J.; Chen, Y. Solution structure of the C-terminal domain of Ku80 suggests important sites for protein-protein interactions. Structure 2004, 12, 495–502.
- Ono, M.; Tucker, P.W.; Capra, J.D. Production and characterization of recombinant human Ku antigen. Nucleic Acids Res. 1994, 22, 3918–3924.
- Mari, P.-O.; Florea, B.I.; Persengiev, S.P.; Verkaik, N.S.; Brüggenwirth, H.T.; Modesti, M.; Giglia-Mari, G.; Bezstarosti, K.; Demmers, J.A.A.; Luider, T.M.; et al. Dynamic assembly of end-joining complexes requires interaction between Ku70/80 and XRCC4. Proc. Natl. Acad. Sci. USA 2006, 103, 18597–18602.
- Uematsu, N.; Weterings, E.; Yano, K.; Morotomi-Yano, K.; Jakob, B.; Taucher-Scholz, G.; Mari, P.O.; van Gent, D.C.; Chen, B.P.; Chen, D.J. Autophosphorylation of DNA-PKCS regulates its dynamics at DNA double-strand breaks. J. Cell Biol. 2007, 177, 219–229.
- Shanbhag, N.M.; Rafalska-Metcalf, I.U.; Balane-Bolivar, C.; Janicki, S.M.; Greenberg, R.A. ATM-Dependent Chromatin Changes Silence Transcription In cis to DNA Double-Strand Breaks. Cell 2010, 141, 970–981.
- Michelini, F.; Pitchiaya, S.; Vitelli, V.; Sharma, S.; Gioia, U.; Pessina, F.; Cabrini, M.; Wang, Y.; Capozzo, I.; Iannelli, F.; et al. Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 2017, 19, 1400–1411.
- Michalik, K.M.; Böttcher, R.; Förstemann, K. A small RNA response at DNA ends in Drosophila. Nucleic Acids Res. 2012, 40, 9596–9603.
- D’Alessandro, G.; Whelan, D.R.; Howard, S.M.; Vitelli, V.; Renaudin, X.; Adamowicz, M.; Iannelli, F.; Jones-Weinert, C.W.; Lee, M.; Matti, V.; et al. BRCA2 controls DNA:RNA hybrid level at DSBs by mediating RNase H2 recruitment. Nat. Commun. 2018, 9, 5376.
- Ohle, C.; Tesorero, R.; Schermann, G.; Dobrev, N.; Sinning, I.; Fischer, T. Transient RNA-DNA Hybrids Are Required for Efficient Double-Strand Break Repair. Cell 2016, 167, 1001–1013.e1007.
- Francia, S.; Michelini, F.; Saxena, A.; Tang, D.; de Hoon, M.; Anelli, V.; Mione, M.; Carninci, P.; d’Adda di Fagagna, F. Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 2012, 488, 231–235.
- Bader, A.S.; Bushell, M. DNA:RNA hybrids form at DNA double-strand breaks in transcriptionally active loci. Cell Death Dis. 2020, 11, 280.
- Trott, D.A.; Porter, A.C.G. Hypothesis: Transcript-templated repair of DNA double-strand breaks. BioEssays 2006, 28, 78–83.
- Lavigne, M.D.; Konstantopoulos, D.; Ntakou-Zamplara, K.Z.; Liakos, A.; Fousteri, M. Global unleashing of transcription elongation waves in response to genotoxic stress restricts somatic mutation rate. Nat. Commun. 2017, 8, 2076.
- Liu, X.; Bushnell, D.A.; Kornberg, R.D. RNA polymerase II transcription: Structure and mechanism. Biochim. Biophys. Acta 2013, 1829, 2–8.
- Becherel, O.J.; Yeo, A.J.; Stellati, A.; Heng, E.Y.; Luff, J.; Suraweera, A.M.; Woods, R.; Fleming, J.; Carrie, D.; McKinney, K.; et al. Senataxin plays an essential role with DNA damage response proteins in meiotic recombination and gene silencing. PLoS Genet. 2013, 9, e1003435.
- Beli, P.; Lukashchuk, N.; Wagner, S.A.; Weinert, B.T.; Olsen, J.V.; Baskcomb, L.; Mann, M.; Jackson, S.P.; Choudhary, C. Proteomic investigations reveal a role for RNA processing factor THRAP3 in the DNA damage response. Mol. Cell 2012, 46, 212–225.
- Jain, A.; Bacolla, A.; Del Mundo, I.M.; Zhao, J.; Wang, G.; Vasquez, K.M. DHX9 helicase is involved in preventing genomic instability induced by alternatively structured DNA in human cells. Nucleic Acids Res. 2013, 41, 10345–10357.
- Cohen, S.; Puget, N.; Lin, Y.-L.; Clouaire, T.; Aguirrebengoa, M.; Rocher, V.; Pasero, P.; Canitrot, Y.; Legube, G. Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat. Commun. 2018, 9, 533.
- Lu, W.-T.; Hawley, B.R.; Skalka, G.L.; Baldock, R.A.; Smith, E.M.; Bader, A.S.; Malewicz, M.; Watts, F.Z.; Wilczynska, A.; Bushell, M. Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nat. Commun. 2018, 9, 532.
- Rawal, C.C.; Zardoni, L.; Di Terlizzi, M.; Galati, E.; Brambati, A.; Lazzaro, F.; Liberi, G.; Pellicioli, A. Senataxin Ortholog Sen1 Limits DNA:RNA Hybrid Accumulation at DNA Double-Strand Breaks to Control End Resection and Repair Fidelity. Cell Rep. 2020, 31, 107603.
- Hegazy, Y.A.; Fernando, C.M.; Tran, E.J. The balancing act of R-loop biology: The good, the bad, and the ugly. J. Biol. Chem. 2020, 295, 905–913.
- Meers, C.; Keskin, H.; Banyai, G.; Mazina, O.; Yang, T.; Gombolay, A.L.; Mukherjee, K.; Kaparos, E.I.; Newnam, G.; Mazin, A.; et al. Genetic Characterization of Three Distinct Mechanisms Supporting RNA-Driven DNA Repair and Modification Reveals Major Role of DNA Polymerase ζ. Mol. Cell 2020, 79, 1037–1050.e1035.
- Mazina, O.M.; Somarowthu, S.; Kadyrova, L.Y.; Baranovskiy, A.G.; Tahirov, T.H.; Kadyrov, F.A.; Mazin, A.V. Replication protein A binds RNA and promotes R-loop formation. J. Biol. Chem. 2020, 295, 14203–14213.
- Keskin, H.; Meers, C.; Storici, F. Transcript RNA supports precise repair of its own DNA gene. RNA Biol. 2016, 13, 157–165.
- Chandramouly, G.; Zhao, J.; McDevitt, S.; Rusanov, T.; Hoang, T.; Borisonnik, N.; Treddinick, T.; Lopezcolorado, F.W.; Kent, T.; Siddique, L.A.; et al. Polθ reverse transcribes RNA and promotes RNA-templated DNA repair. Sci. Adv. 2021, 7, eabf1771.
- Ferenczi, A.; Pyott, D.E.; Xipnitou, A.; Molnar, A. Efficient targeted DNA editing and replacement in Chlamydomonas reinhardtii using Cpf1 ribonucleoproteins and single-stranded DNA. Proc. Natl. Acad. Sci. USA 2017, 114, 13567–13572.
- Yang, Y.G.; Lindahl, T.; Barnes, D.E. Trex1 exonuclease degrades ssDNA to prevent chronic checkpoint activation and autoimmune disease. Cell 2007, 131, 873–886.
- Lanza, D.G.; Gaspero, A.; Lorenzo, I.; Liao, L.; Zheng, P.; Wang, Y.; Deng, Y.; Cheng, C.; Zhang, C.; Seavitt, J.R.; et al. Comparative analysis of single-stranded DNA donors to generate conditional null mouse alleles. BMC Biol. 2018, 16, 69.
- Yang, Y.G.; Qi, Y. RNA-directed repair of DNA double-strand breaks. DNA Repair 2015, 32, 82–85.
- Shen, Y.; Nandi, P.; Taylor, M.B.; Stuckey, S.; Bhadsavle, H.P.; Weiss, B.; Storici, F. RNA-driven genetic changes in bacteria and in human cells. Mutat. Res. Fundam. Mol. Mech. Mutagenesis 2011, 717, 91–98.
More
