The Y chromosome is one of the sex chromosomes found in males of animals of different taxa, including insects and mammals. Among all chromosomes, the Y chromosome is characterized by a unique chromatin landscape undergoing dynamic evolutionary change. Being entirely heterochromatic, the Y chromosome as a rule preserves few functional genes, but is enriched in tandem repeats and transposons. Due to difficulties in the assembly of the highly repetitive Y chromosome sequence, deep analyses of Y chromosome evolution, structure, and functions are limited to a few species, one of them being Drosophila melanogaster. Here researchers survey comparative evolutionary history of the fly and human Y chromosomes, and functions of Y-linked piRNA clusters ensuring sex-specific piRNA silencing.
The Y chromosome is one of the sex chromosomes found in males of animals of different taxa, including insects and mammals. Among all chromosomes, the Y chromosome is characterized by a unique chromatin landscape undergoing dynamic evolutionary change. Being entirely heterochromatic, the Y chromosome as a rule preserves few functional genes, but is enriched in tandem repeats and transposons. Due to difficulties in the assembly of the highly repetitive Y chromosome sequence, deep analyses of Y chromosome evolution, structure, and functions are limited to a few species, one of them being Drosophila melanogaster. Here we survey comparative evolutionary history of the fly and human Y chromosomes, and functions of Y-linked piRNA clusters ensuring sex-specific piRNA silencing.
- Drosophila
- Y chromosome
- piRNA pathway
- rDNA
- intron gigantism
- azoospermia
- transposable elements
1. Introduction
2. Comparative Evolutionary History of the Fly and Human Y Chromosomes
2.1. Y Chromosome Differentiation and Functions in Flies
The Y chromosome is a sex chromosome found in males of different groups of animals, including mammals and Diptera. Whereas in mammals the development of an organism according to the male type is determined by the presence of a functional Y chromosome, in Drosophila, sex is determined by the ratio of the number of X chromosomes to the number of the autosomes: normally, the presence of two X chromosomes triggers development according to the female type, and one—according to the male type [8][9]. Thus, individuals with the XXY genotype are female in flies and male in mammals, while X0 are female in mammals and male in flies. In Drosophila with the X0 karyotype, there are no severe structural or functional body disorders, except for male sterility [6]. Fly Y chromosome is not involved in sex determination. In Diptera, the Y chromosomes arose from the autosomes repeatedly (Figure 2a), which provides good material for studying parallel processes of convergent evolutionary development [10][11][12][13]. Relatively young, newly formed Y chromosomes of flies, being formed from tens of thousands of years to a few million years ago, maintain the structure and genes of the ancestral autosome, while most of the genes on the old fly Y chromosomes have been acquired subsequently due to a transfer from the autosomes or the X chromosome. Old Y chromosomes such as in D. melanogaster that presumably have been persisted for long time (several decades of millions of years) are often highly heterochromatic, contain a large amount of repetitive DNA, and their genes undergo degeneration [10][12].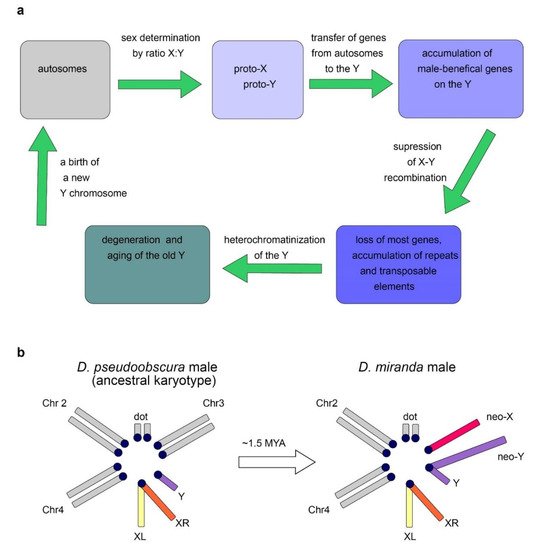
2.2. Origin of the Y Chromosome in Mammals and Sex Determination
In many animals, sex is determined by a pair of heteromorphic X and Y chromosomes. According to modern concepts, sex chromosomes originate from an ancestral pair of autosomes, one of which acquires a sex-specific gene, which starts the process of differentiation of the sex chromosomes. In mammals, this event occurred only once in the common ancestor of marsupials and placentals prior to their splitting, about 160–180 MYA [19][20][21][22]. The proto-Y chromosome of all mammals (from kangaroo to human) arose from a single autosome in which one of the alleles of the SOX3 gene, as a result of a mutation, became the sex-determining gene SRY [23][24]. However, the product of the sex determination gene only provides a switch, triggering a certain pathway of development. Unique evolutionary forces facilitated the selection and accumulation of male-beneficial mutations around the SRY locus, and the linkage between them was supported by selective pressure to avoid crossing over between the proto-Y and proto-X [25]. As a rule, if the dominant allele causes the development of a male, then the chromosome in which it is located becomes the Y chromosome (and its homolog is called the X). In birds, males are the homogametic sex (ZZ) and females are the heterogametic (ZW) [21][26].2.3. Evolutionary Factors and Forces Determining the Structure and Functional Specialization of the Y Chromosome
The loss of recombination leads to the inefficiency of natural selection and causes the ensuing accumulation of Y-linked loss-of-function mutations, chromosome-wide gene decay, and amplification of repetitive DNAs [27][28][29][30]. In parallel to the loss of genes, Y chromosomes have accumulated large amounts of DNA repeats, and the D. melanogaster old Y chromosome mainly consists of heterochromatin (Figure 2a) [4][21]. Despite the human Y chromosome having undergone a rapid decay early in evolution, its massive degeneration then dramatically stopped. Genes that remained intact currently show remarkable stability, and no human Y-linked genes have been lost during the last 44 million years [22][31]. The maintenance of human Y-linked genes is mainly associated with two functional categories: genes essential for male reproductive functions and dosage-sensitive ubiquitous housekeepers [32]. Studies of males with Y deletions have allowed researchers to identify three ‘azoospermia factor’ (AZF) regions, AZFa, AZFb, and AZFc, and partially map within them the genes essential for spermatogenesis [33]. The AZFa deletions affecting the DBY gene cause the most severe azoospermia phenotype, exhibiting a complete loss of testis germline cells accompanied by the maintenance of somatic Sertoli cells (the so-called Sertoli Cell-Only Syndrome; SCOS) [34][35][36]. As in fruit flies, mammalian Y chromosomes also exhibit gene amplification, with the amplicon structures predominantly containing genes with testis-specific functions [37][38][39][40]. Due to the presence of repeating structures, local intra-chromosomal gene conversion is possible, as well as intra- and inter-chromatid exchange. These mechanisms partially compensate for the lack of recombination with the X chromosome by eliminating harmful mutations. At the same time, inter-chromatid recombination can in some cases lead to the formation of isodicentric chromosomes formed by homologous crossing over between opposing arms of palindromes on sister chromatids [41]. The loss of the ability to recombine plays a key role in establishing the structure of the Y chromosome [30][42]. Mutations that prevent recombination between proto-X and proto-Y, such as inversions, deletions, or accumulation of repeats, are supported by selection. Reducing the ability of recombination with the homologous X chromosome dramatically accelerated the evolution of the Y chromosome preventing the elimination of emerging mutations via crossing over, while the X chromosome has retained the ability to cross over in the homogametic sex. This led to the degeneration of most of the original Y-chromosomal genes, and multiple deletions caused a significant size decrease with a relative increase in the proportion of non-coding heterochromatic regions. The rapid evolutionary degeneration of the Y chromosomes, typical in a wide range of species, leads to the hypothesis that in the future the human Y chromosome may disappear altogether. This hypothesis is based not only on extrapolation, but is also indirectly supported by precedents in the evolution of some species including multiple fishes, reptiles, grasshoppers, cockroaches, and dragonflies [43][44][45]. However, other researchers claim that human Y degeneration stopped millions of years ago and currently nothing threatens Y chromosome survival [46].2.4. Dosage Compensation System Contributes to Y-Linked Gene Maintenance
As a rule, a single gene copy appears to be enough to provide development and life-cycle maintenance of diploid animals; however, a small cohort of genes exhibits a high sensitivity in case of decreased gene dosage. This phenomenon is known as haploinsufficiency, and it is associated with many developmental disorders in human [47][48][49]. In male flies, the genes of the only X chromosome are overactivated in somatic tissues, eliminating the problem of haploinsufficiency and potentially lethal imbalance between the X and autosome transcriptional level in the two sexes. In contrast, in female mammals, inactivation of one of the two X chromosomes occurs. However, according to various estimates and in distinct types of human cells, 20–30% of genes of inactive X chromosome escape the inactivation [50][51]. In mammals, haploinsufficient Y-chromosomal genes have X-chromosomal homologues that avoid inactivation during dosage compensation in females, which indicates the need for their expression on both sex chromosomes to ensure normal functions in the body. Thus, in males, these dosage-sensitive genes cannot disappear from the Y chromosome without negative consequences, and they can survive under selective pressure [31][32][50][52]. Strict dosage requirements for sex-linked genes are demonstrated in the case of Turner syndrome (exhibiting X0 karyotype or mosaicism) and Klinefelter syndrome (XXY), since such genes have been haploinsufficient or overexpressed, respectively, in these karyotypes [51]. Turner syndrome is a genetic condition caused by complete or partial loss of the second sex chromosome in human [53][54]. Studies of manifestations of this syndrome indicate that the functions of the Y chromosome consist not only of ensuring the normal functioning of the male reproductive system. Due to the absence of the SRY gene, which is the key to triggering male-type development, patients with this syndrome are exclusively female, with multiple body disorders and cognitive impairment [54]. Individuals with Klinefelter syndrome are infertile as a result of excess gene dosage of X escape genes, and abnormal meiotic pairing of the sex chromosomes. An atypical number of X or Y chromosomes (XXY, XXX, or X) contributes to spatial chromosome conformation changes and leads to disruption of DNA methylation patterns of autosomal genes, causing distinct disease phenotypes: mental illness, cancer, and disrupted fertility [51].2.5. Convergent Nature of the Evolution of Y Chromosomes
Despite their independent evolutionary origins in different species, Y chromosomes in species with heterogametic males have a number of similar features: they are usually smaller than X chromosomes, contain significantly fewer genes, most of which are related to the male reproductive system, and also have a relatively large number of repeats and significant areas occupied by heterochromatin. It has been proposed that such convergent evolution is due to the similar nature of the selection pressure. Another common feature—the acquisition of repetitive sequences and the loss of most of the original genes—is associated with accelerated Y evolution due to the loss of recombination with the X chromosome [27]. The difference between the evolution of the Y chromosome in mammals and Diptera is mainly that in Diptera the acquisition of new genes often significantly prevails over the loss of the original ones; although, both processes take place in both groups. Presumably due to slower changes in mammals, the evolutionary processes have not yet reached the point where the Y chromosome has lost all homology with the X chromosome.3. Current Undestanding of Drosophila Y Chromosome Contribution in piRNA Biogenesis and Functioning of piRNA-Clusters
3.1. Brief Description of the piRNA System
The piRNA pathway provides both innate and adaptive immune system defense against the activity of transposable elements (TEs) leading to the protection of genome integrity in germinal tissues. It also participates in the maintenance of germline stem cells, regulation of protein-coding gene expression, the establishment of embryonic patterning (in Diptera), and transgenerational epigenetic inheritance [55][56][57]. Small non-coding piRNAs 23-35 nt in length associated with proteins of the PIWI subfamily are present in animals from fungi to humans [58][59][60]. piRNAs are generated from piRNA clusters, which are long precursors that are transcribed from heterochromatic regions containing fragments of transposons. piRNA precursors are processed to generate small piRNAs in perinuclear nuage granules (Figure 3) [61][62][63][64].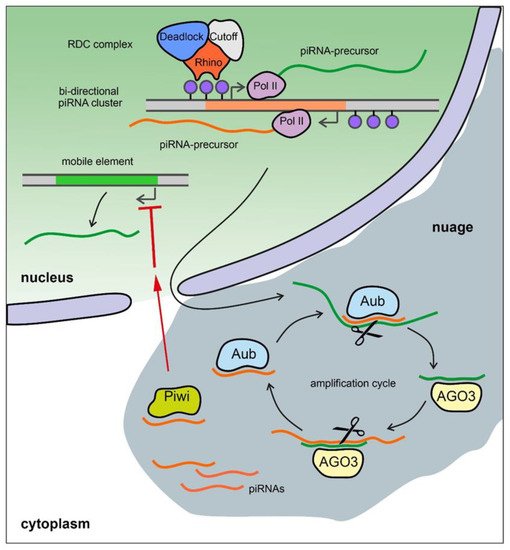
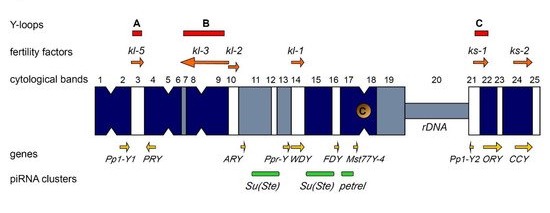
3.2. The Y Chromosome as a Major piRNA-Producing Genomic Region in the Fly Testes
The piRNA system in D. melanogaster exhibits a strong sexual dimorphism. TE-mapping piRNAs are known as the most abundant class of piRNAs in the ovaries, whereas only about 40% of piRNAs map to TEs in the testes, and the largest cohort of piRNAs map to protein-coding genes [7][65]. In the testes of Drosophila, almost half of all piRNAs originate from the piRNA clusters located on the Y chromosome (Figure 1) [7]. The largest number of piRNAs is generated from the Y-linked Suppressor of Stellate (Su(Ste)) repeats directed to silencing of the homologous tandem Stellate genes residing on the X chromosome [65][66][67]. The number of Su(Ste) repeats comprises more than 500 tandemly ordered copies residing in two cytolocations on the Y (Figure 1) [2][4][7]. The insertion of the defective transposon hoppel into the promoter is responsible for the initiation of antisense transcription of Su(Ste) repeats and their acquisition of piRNA cluster functions [66]. Stellate derepression in the case of deletion of most of Su(Ste) repeats or disruption of the piRNA system leads to the accumulation of needle-like protein aggregates in spermatocytes, disturbances of meiosis, and, as a result, a decrease in male fertility [66][68]. The Stellate/Su(Ste) system is species- and sex-specific for D. melanogaster. It was is shown that Stellate genes participate in male hybrid sterility of F1 progeny of crosses between D. melanogaster females and males of closely related D. mauritiana. The hybrid males possess maternal X-linked Stellate genes, but their paternal Y chromosome does not contain Su(Ste) repeats and the corresponding piRNAs are not generated. Derepression of Stellates in the testes of hybrid males leads to a meiotic catastrophe and complete sterility [65][68]. The Y chromosome of D. melanogaster also contains the petrel locus (Figure 1), which is a source of multiple piRNAs highly complementary to pirate/CG12717 gene, providing strong testis-specific silencing of this gene [7]. However, the functional significance of the repression of pirate,encoding a SUMO-isopeptidase, in the testes remains unclear to date. It appears that both in the cases of the Stellate/Su(Ste) and pirate/petrel pairs, their current evolutionary relationships are initially based on parallel acquisition or co-amplification of homologous genes on the sex chromosomes. On the whole, the mechanism of determination of genomic regions as piRNA clusters is poorly resolved.3.3. The Y Chromosome in Other Species as a Source of piRNAs
The suppression of genes harmful for spermatogenesis appears to be one of the main functions of piRNAs originating from the Y chromosome of D. melanogaster. In mouse testes, novel polyadenylated non-coding RNAs called Pirmy and Pirmy-like transcribed from the long arm of the Y chromosome have recently been discovered [69]. Morphology- and sperm motility-related abnormalities have been found in two strains of Y-deleted mice with disrupted expression of Pirmy and Pirmy-like RNAs. The Pirmy and Pirmy-like RNAs serve as sources of piRNAs that are complementary to 5′- and 3′-UTRs of several autosomal genes, that presumably contribute to fertility and sex ratio maintenance in the progeny. The proteins expressed from these autosomal genes are up-regulated in the sperm of Y-deleted mice and appear to be responsible for the disruption of sperm morphology and motility [69]. In Bombyx mori, females are the heterogametic sex (ZW), and the W chromosome is heterochromatinized and consists almost entirely of transposon sequences. piRNA from the Fem locus on the W chromosome functions as a suppressor of the Masc gene, which regulates sex-specific splicing of the doublesex (dsx) gene, which is necessary for sex determination in many insects [70]. Thus, small piRNAs from the Y or W chromosome can potentially be involved in sex determination, the resolution of intragenomic conflicts, reproductive isolation, and the regulation of gene expression for ensuring spermatogenesis [7][16][65][69][70][71]. Y-linked piRNA clusters and their functions in humans remain poorly understood [72][73]. High-throughput sequencing of piRNAs from three human adult testis samples and subsequent data analysis have revealed 28 putative piRNA-cluster candidate regions on the Y [74]. However, among them, only one uni-directional cluster contains a significant number of mapped piRNAs (45.4 rpkm). This locus includes remnants of SINE, LINE, and LTR TEs, and has a highly homologous region of the same size on the X chromosome. Due to the high level of heterochromatinization and a large number of repetitive elements, the human Y chromosome is not perfectly assembled, and data about piRNA clusters are not complete.4. Conclusions and Pperspectives
In most heterosexual eukaryotes Y chromosomes are maintained and perform various essential functions. These include sex determination, ensuring male fertility; correct segregation of meiotic chromosomes; epigenetic regulation of harmful elements; contribution to interspecies hybrid sterility; and other responsibilities. Studies of model organisms, Drosophila and mice, have fundamental significance for uncovering the shared properties of Y chromosomes of multiple species. The convergent nature of evolution of the Y chromosome allows researchers to consider that the data obtained in model organisms can be useful to a certain extent for the prediction of the human Y chromosome behavior in the future, as well as in understanding how the specific structure of this chromosome reflects its functions in normal and pathological conditions.References
- Bonaccorsi, S.; Lohe, A. Fine mapping of satellite DNA sequences along the Y chromosome of Drosophila melanogaster: Relationships between satellite sequences and fertility factors. Genetics 1991, 129, 177–189.
- Hoskins, R.A.; Carlson, J.W.; Wan, K.H.; Park, S.; Mendez, I.; Galle, S.E.; Booth, B.W.; Pfeiffer, B.D.; George, R.A.; Svirskas, R.; et al. The Release 6 reference sequence of the Drosophila melanogaster genome. Genome Res. 2015, 25, 445–458.
- Carvalho, A.B.; Vicoso, B.; Russo, C.A.; Swenor, B.; Clark, A.G. Birth of a new gene on the Y chromosome of Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 2015, 112, 12450–12455.
- Chang, C.H.; Larracuente, A.M. Heterochromatin-Enriched Assemblies Reveal the Sequence and Organization of the Drosophila melanogaster Y Chromosome. Genetics 2019, 211, 333–348.
- Chang, C.H.; Gregory, L.E.; Gordon, K.E.; Meiklejohn, C.D.; Larracuente, A.M. Unique structure and positive selection promote the rapid divergence of Drosophila Y chromosomes. eLife 2022, 11, e75795.
- Piergentili, R. Multiple roles of the Y chromosome in the biology of Drosophila melanogaster. Sci. World J. 2010, 10, 1749–1767.
- Chen, P.; Kotov, A.A.; Godneeva, B.K.; Bazylev, S.S.; Olenina, L.V.; Aravin, A.A. piRNA-mediated gene regulation and adaptation to sex-specific transposon expression in D. melanogaster male germline. Genes Dev. 2021, 35, 914–935.
- Salz, H.K.; Erickson, J.W. Sex determination in Drosophila: The view from the top. Fly 2010, 4, 60–70.
- Bachtrog, D.; Mank, J.E.; Peichel, C.L.; Kirkpatrick, M.; Otto, S.P.; Ashman, T.L.; Hahn, M.W.; Kitano, J.; Mayrose, I.; Ming, R.; et al. Tree of Sex Consortium. Sex determination: Why so many ways of doing it? PLoS Biol. 2014, 12, e1001899.
- Carvalho, A.B.; Koerich, L.B.; Clark, A.G. Origin and evolution of Y chromosomes: Drosophila tales. Trends Genet. 2009, 25, 270–277.
- Vicoso, B.; Bachtrog, D. Numerous transitions of sex chromosomes in Diptera. PLoS Biol. 2015, 13, e1002078.
- Mahajan, S.; Bachtrog, D. Convergent evolution of Y chromosome gene content in flies. Nat. Commun. 2017, 8, 785.
- Koerich, L.B.; Wang, X.; Clark, A.G.; Carvalho, A.B. Low conservation of gene content in the Drosophila Y chromosome. Nature 2008, 456, 949–951.
- Bachtrog, D.; Mahajan, S.; Bracewell, R. Massive gene amplification on a recently formed Drosophila Y chromosome. Nat. Ecol. Evol. 2019, 3, 1587–1597.
- Bachtrog, D.; Charlesworth, B. Reduced adaptation of a non-recombining neo-Y chromosome. Nature 2002, 416, 323–326.
- Bachtrog, D. The Y Chromosome as a Battleground for Intragenomic Conflict. Trends Genet. 2020, 36, 510–522.
- Carvalho, A.B.; Clark, A.G. Y chromosome of D. pseudoobscura is not homologous to the ancestral Drosophila Y. Science 2005, 307, 108–110.
- Ricchio, J.; Uno, F.; Carvalho, A.B. New Genes in the Drosophila Y Chromosome: Lessons from D. willistoni. Genes 2021, 12, 1815.
- Ohno, S. Sex Chromosomes and Sex-Linked Genes; Monographs on Endocrinology; Springer: Berlin/Heidelberg, Germany, 1967.
- Potrzebowski, L.; Vinckenbosch, N.; Marques, A.C.; Chalmel, F.; Jégou, B.; Kaessmann, H. Chromosomal gene movements reflect the recent origin and biology of therian sex chromosomes. PLoS Biol. 2008, 6, e80.
- Veyrunes, F.; Waters, P.D.; Miethke, P.; Rens, W.; McMillan, D.; Alsop, A.E.; Grützner, F.; Deakin, J.E.; Whittington, C.M.; Schatzkamer, K.; et al. Bird-like sex chromosomes of platypus imply recent origin of mammal sex chromosomes. Genome Res. 2008, 18, 965–973.
- Subrini, J.; Turner, J. Y chromosome functions in mammalian spermatogenesis. eLife 2021, 10, e67345.
- Foster, J.W.; Graves, J.A. An SRY-related sequence on the marsupial x chromosome: Implications for the evolution of the mammalian testis-determining gene. Proc. Natl. Acad. Sci. USA 1994, 91, 1927–1931.
- Graves, J.A. Evolution of the mammalian Y chromosome and sex-determining genes. J. Exp. Zool. 1998, 281, 472–481.
- Larson, E.L.; Kopania, E.E.K.; Good, J.M. Spermatogenesis and the evolution of mammalian sex chromosomes. Trends Genet. 2018, 34, 722–732.
- Ioannidis, J.; Taylor, G.; Zhao, D.; Liu, L.; Idoko-Akoh, A.; Gong, D.; Lovell-Badge, R.; Guioli, S.; McGrew, M.J.; Clinton, M. Primary sex determination in birds depends on DMRT1 dosage, but gonadal sex does not determine adult secondary sex characteristics. Proc. Natl. Acad. Sci. USA 2021, 118, e2020909118.
- Rice, W.R. The accumulation of sexually antagonistic genes as a selective agent promoting the evolution of reduced recombination between primitive sex chromosomes. Evolution 1987, 41, 911–914.
- Rice, W.R. Genetic hitchhiking and the evolution of reduced genetic activity of the Y sex chromosome. Genetics 1987, 116, 161–167.
- Bachtrog, D. Y-chromosome evolution: Emerging insights into processes of Y-chromosome degeneration. Nat. Rev. Genet. 2013, 14, 113–124.
- Charlesworth, B.; Charlesworth, D. The degeneration of Y chromosomes. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2000, 355, 1563–1572.
- Bellott, D.W.; Hughes, J.F.; Skaletsky, H.; Brown, L.G.; Pyntikova, T.; Cho, T.J.; Koutseva, N.; Zaghlul, S.; Graves, T.; Rock, S.; et al. Mammalian Y chromosomes retain widely expressed dosage-sensitive regulators. Nature 2014, 508, 494–499.
- Lahn, B.T.; Page, D.C. Functional coherence of the human Y chromosome. Science 1997, 278, 675–680.
- Vogt, P.H. AZF deletions and Y chromosomal haplogroups: History and update based on sequence. Hum. Reprod. Update 2005, 11, 319–336.
- Kamp, C.; Huellen, K.; Fernandes, S.; Sousa, M.; Schlegel, P.N.; Mielnik, A.; Kleiman, S.; Yavetz, H.; Krause, W.; Küpker, W.; et al. High deletion frequency of the complete AZFa sequence in men with Sertoli-cell-only syndrome. Mol. Hum. Reprod. 2001, 7, 987–994.
- Lardone, M.C.; Parodi, D.A.; Valdevenito, R.; Ebensperger, M.; Piottante, A.; Madariaga, M.; Smith, R.; Pommer, R.; Zambrano, N.; Castro, A. Quantification of DDX3Y, RBMY1, DAZ and TSPY mRNAs in testes of patients with severe impairment of spermatogenesis. Mol. Hum. Reprod. 2007, 13, 705–712.
- Kotov, A.A.; Olenkina, O.M.; Godneeva, B.K.; Adashev, V.E.; Olenina, L.V. Progress in understanding the molecular functions of DDX3Y (DBY) in male germ cell development and maintenance. Biosci. Trends 2017, 11, 46–53.
- Bhowmick, B.K.; Satta, Y.; Takahata, N. The origin and evolution of human ampliconic gene families and ampliconic structure. Genome Res. 2007, 17, 441–450.
- Skaletsky, H.; Kuroda-Kawaguchi, T.; Minx, P.J.; Cordum, H.S.; Hillier, L.; Brown, L.G.; Repping, S.; Pyntikova, T.; Ali, J.; Bieri, T.; et al. The male-specific region of the human Y chromosome is a mosaic of discrete sequence classes. Nature 2003, 423, 825–837.
- Betrán, E.; Demuth, J.P.; Williford, A. Why chromosome palindromes? Int. J. Evol. Biol. 2012, 2012, 207958.
- Bonito, M.; D’Atanasio, E.; Ravasini, F.; Cariati, S.; Finocchio, A.; Novelletto, A.; Trombetta, B.; Cruciani, F. New insights into the evolution of human Y chromosome palindromes through mutation and gene conversion. Hum. Mol. Genet. 2021, 30, 2272–2285.
- Lange, J.; Skaletsky, H.; van Daalen, S.K.; Embry, S.L.; Korver, C.M.; Brown, L.G.; Oates, R.D.; Silber, S.; Repping, S.; Page, D.C. Isodicentric Y chromosomes and sex disorders as byproducts of homologous recombination that maintains palindromes. Cell 2009, 138, 855–869.
- Hughes, J.F.; Page, D.C. The Biology and Evolution of Mammalian Y Chromosomes. Annu. Rev. Genet. 2015, 49, 507–527.
- Traut, W.; Winking, H. Meiotic chromosomes and stages of sex chromosome evolution in fish: Zebrafish, platyfish and guppy. Chromosome Res. 2001, 9, 659–672.
- Graves, J.A. Sex chromosome specialization and degeneration in mammals. Cell 2006, 124, 901–914.
- Blackmon, H.; Ross, L.; Bachtrog, D. Sex Determination, Sex Chromosomes, and Karyotype Evolution in Insects. J. Hered. 2017, 108, 78–93.
- Griffin, D.K. Is the Y chromosome disappearing?—Both sides of the argument. Chromosome Res. 2012, 20, 35–45.
- Fisher, E.; Scambler, P. Human haploinsufficiency—One for sorrow, two for joy. Nat. Genet. 1994, 7, 5–7.
- Johnson, A.F.; Nguyen, H.T.; Veitia, R.A. Causes and effects of haploinsufficiency. Biol. Rev. 2019, 94, 1774–1785.
- Zug, R. Developmental disorders caused by haploinsufficiency of transcriptional regulators: A perspective based on cell fate determination. Biol. Open. 2022, 11, bio058896.
- Carrel, L.; Brown, C.J. When the Lyon(ized chromosome) roars: Ongoing expression from an inactive X chromosome. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2017, 372, 20160355.
- Fang, H.; Disteche, C.M.; Berletch, J.B. X Inactivation and Escape: Epigenetic and Structural Features. Front. Cell Dev. Biol. 2019, 7, 219.
- Cortez, D.; Marin, R.; Toledo-Flores, D.; Froidevaux, L.; Liechti, A.; Waters, P.D.; Grützner, F.; Kaessmann, H. Origins and functional evolution of Y chromosomes across mammals. Nature 2014, 508, 488–493.
- Hook, E.B.; Warburton, D. Turner syndrome revisited: Review of new data supports the hypothesis that all viable 45, X cases are cryptic mosaics with a rescue cell line, implying an origin by mitotic loss. Hum. Genet. 2014, 133, 417–424.
- Gravholt, C.H.; Viuff, M.H.; Brun, S.; Stochholm, K.; Andersen, N.H. Turner syndrome: Mechanisms and management. Nat. Rev. Endocrinol. 2019, 15, 601–614.
- Le Thomas, A.; Stuwe, E.; Li, S.; Du, J.; Marinov, G.; Rozhkov, N.; Chen, Y.C.; Luo, Y.; Sachidanandam, R.; Toth, K.F.; et al. Transgenerationally inherited piRNAs trigger piRNA biogenesis by changing the chromatin of piRNA clusters and inducing precursor processing. Genes Dev. 2014, 28, 1667–1680.
- Ozata, D.M.; Gainetdinov, I.; Zoch, A.; O’Carroll, D.; Zamore, P.D. PIWI-interacting RNAs: Small RNAs with big functions. Nat. Rev. Genet. 2019, 20, 89–108.
- Ramat, A.; Simonelig, M. Functions of PIWI Proteins in Gene Regulation: New Arrows Added to the piRNA Quiver. Trends Genet. 2021, 37, 188–200.
- Aravin, A.A.; Hannon, G.J.; Brennecke, J. The Piwi-piRNA pathway provides an adaptive defense in the transposon arms race. Science 2007, 318, 761–764.
- Malone, C.D.; Brennecke, J.; Dus, M.; Stark, A.; McCombie, W.R.; Sachidanandam, R.; Hannon, G.J. Specialized piRNA pathways act in germline and somatic tissues of the Drosophila ovary. Cell 2009, 137, 522–535.
- Gainetdinov, I.; Colpan, C.; Arif, A.; Cecchini, K.; Zamore, P.D. A Single Mechanism of Biogenesis, Initiated and Directed by PIWI Proteins, Explains piRNA Production in Most Animals. Mol. Cell 2018, 71, 775–790.e5.
- Brennecke, J.; Aravin, A.A.; Stark, A.; Dus, M.; Kellis, M.; Sachidanandam, R.; Hannon, G.J. Discrete small RNA-generating loci as master regulators of transposon activity in Drosophila. Cell 2007, 128, 1089–1103.
- Mohn, F.; Sienski, G.; Handler, D.; Brennecke, J. The Rhino-Deadlock-Cutoff Complex Licenses Noncanonical Transcription of Dual-Strand piRNA Clusters in Drosophila. Cell 2014, 157, 1364–1379.
- Chen, Y.A.; Stuwe, E.; Luo, Y.; Ninova, M.; Le Thomas, A.; Rozhavskaya, E.; Li, S.; Vempati, S.; Laver, J.D.; Patel, D.J.; et al. Cutoff Suppresses RNA Polymerase II Termination to Ensure Expression of piRNA Precursors. Mol. Cell. 2016, 63, 97–109.
- Zhang, Z.; Wang, J.; Schultz, N.; Zhang, F.; Parhad, S.S.; Tu, S.; Vreven, T.; Zamore, P.D.; Weng, Z.; Theurkauf, W.E. The HP1 homolog rhino anchors a nuclear complex that suppresses piRNA precursor splicing. Cell 2014, 157, 1353–1363.
- Kotov, A.A.; Adashev, V.E.; Godneeva, B.K.; Ninova, M.; Shatskikh, A.S.; Bazylev, S.S.; Aravin, A.A.; Olenina, L.V. piRNA silencing contributes to interspecies hybrid sterility and reproductive isolation in Drosophila melanogaster. Nucleic Acids Res. 2019, 47, 4255–4271.
- Aravin, A.A.; Naumova, N.M.; Tulin, A.V.; Vagin, V.V.; Rozovsky, Y.M.; Gvozdev, V.A. Double-stranded RNA-mediated silencing of genomic tandem repeats and transposable elements in the D. melanogaster germline. Curr. Biol. 2001, 11, 1017–1027.
- Vagin, V.V.; Sigova, A.; Li, C.; Seitz, H.; Gvozdev, V.; Zamore, P.D. A distinct small RNA pathway silences selfish genetic elements in the germline. Science 2006, 313, 320–324.
- Adashev, V.E.; Kotov, A.A.; Bazylev, S.S.; Shatskikh, A.S.; Aravin, A.A.; Olenina, L.V. Stellate Genes and the piRNA Pathway in Speciation and Reproductive Isolation of Drosophila melanogaster. Front. Genet. 2021, 11, 610665.
- Reddy, H.M.; Bhattacharya, R.; Tiwari, S.; Mishra, K.; Annapurna, P.; Jehan, Z.; Praveena, N.M.; Alex, J.L.; Dhople, V.M.; Singh, L.; et al. Y chromosomal noncoding RNAs regulate autosomal gene expression via piRNAs in mouse testis. BMC Biol. 2021, 19, 198.
- Kiuchi, T.; Koga, H.; Kawamoto, M.; Shoji, K.; Sakai, H.; Arai, Y.; Ishihara, G.; Kawaoka, S.; Sugano, S.; Shimada, T.; et al. A single female-specific piRNA is the primary determiner of sex in the silkworm. Nature 2014, 509, 633–636.
- Ellison, C.; Bachtrog, D. Recurrent gene co-amplification on Drosophila X and Y chromosomes. PLoS Genet. 2019, 15, e1008251.
- Sarkar, A.; Maji, R.K.; Saha, S.; Ghosh, Z. piRNAQuest: Searching the piRNAome for silencers. BMC Genom. 2014, 15, 555.
- Özata, D.M.; Yu, T.; Mou, H.; Gainetdinov, I.; Colpan, C.; Cecchini, K.; Kaymaz, Y.; Wu, P.H.; Fan, K.; Kucukural, A.; et al. Evolutionarily conserved pachytene piRNA loci are highly divergent among modern humans. Nat. Ecol. Evol. 2020, 4, 156–168.
- Ha, H.; Song, J.; Wang, S.; Kapusta, A.; Feschotte, C.; Chen, K.C.; Xing, J. A comprehensive analysis of piRNAs from adult human testis and their relationship with genes and mobile elements. BMC Genom. 2014, 15, 545.
