You're using an outdated browser. Please upgrade to a modern browser for the best experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Peter Pelka.
The Never in mitosis gene A (NIMA) family of serine/threonine kinases is a diverse group of protein kinases implicated in a wide variety of cellular processes, including cilia regulation, microtubule dynamics, mitotic processes, cell growth, and DNA damage response. The founding member of this family was initially identified in Aspergillus and was found to play important roles in mitosis and cell division. The yeast family has one member each, Fin1p in fission yeast and Kin3p in budding yeast, also with functions in mitotic processes, but, overall, these are poorly studied kinases.
- NIMA kinases
- Nek
- mitosis
- cell cycle
1. Introduction and Aspergillus NIMA
The NIMA family of kinases is a relatively poorly understood family of serine/threonine kinases. The prototype, NIMA (NIMA—never in mitosis, gene A), was first identified in Aspergillus as a protein responsible for regulation of mitotic progression [1]. In this sentudry, Morris screened temperature-sensitive mutants defective in nuclear division, septation, or distribution of nuclei along the mycelium and classified cell cycle mutants into two categories: those that arrested before entry into mitosis based on a co-ordinate drop in both spindle and chromosome mitotic indices are nim mutants for never in mitosis, and those that arrested during mitosis based on a rise in the same indices are bim mutants for blocked in mitosis. The nim mutants included a variety of cell cycle regulators, such as cyclin B, Cdc25, and DNA polymerase. The bim mutants included genes for spindle motors and components of the anaphase promoting complex. In his screens, Morris identified four alleles of a gene that he denoted nimA [2]. The nimA mutants were found to arrest in late G2 phase, with duplicated spindle pole bodies (the fungal equivalent of the centrosomes; [3]), indicating that NIMA not only affects chromosomal condensation but also other events of mitosis such as spindle formation. Soon thereafter, the gene for nimA was cloned and identified as a serine/threonine protein kinase called NIMA [4]. Overexpression of NIMA leads to a mitotic-like state, with chromosomes that have condensed and the appearance of aberrant spindles. Chromosomes were also observed to condense if cells were arrested in S phase prior to induction of overexpression of NIMA [5]. The activity of NIMA is tightly controlled by several different mechanisms that ensure the protein is active only during G2/M transition [4,6,7][4][6][7]. Mechanisms controlling the activity of the protein include transcriptional control of the nimA locus, phosphorylation at several different sites by Cdc2/Cyclin B complex, dephosphorylation by Cdc25, and loss of protein stability via proteolytic degradation.
Cdc2 activation and its nuclear localization is a prerequisite for entry into mitosis and, in most cases, once Cdc2 has been activated, mitosis will occur. This does not happen in nimA5 mutants [8]. In the nimA5 mutant, Cdc2 is fully active but no mitotic entry occurs, suggesting that a functional NIMA is required for G2/M transition. Activation of NIMA is dependent upon the activity of Cdc2 [6], possibly being directly phosphorylated and activated by Cdc2. In Aspergillus, the nuclear membrane is not broken down during mitosis and Cyclin B/Cdc2 complexes are excluded from the nucleus in nimA mutants [9]. The NIMA protein is nuclear and is associated with chromosomes, spindles, and spindle pole bodies [10]. Cytoplasmic localization of Cdc2 will prevent proper activation of downstream Cdc2 targets that are nuclear. It is likely that NIMA promotes nuclear localization of the Cyclin B/Cdc2 complex, and this is supported by experimental evidence in which a mutation that suppresses the nimA1 allele was found to affect sonA, a homolog of the Gle2/Rae1 nuclear transporter [9]. Another mutation that suppresses nimA1 allele is sonB. The sonB gene encodes a homolog of the human NUP98/NUP96 precursor, which is cleaved to form a mature nucleoporin that contains the GLEBS domain that binds SONA [11]. NIMA and SONA/SONB interact and likely regulate the nuclear entry of mitotic factors, including NIMA itself, that promote entry and progression through mitosis in Aspergillus. Both SONA and SONB contain multiple NIMA consensus phosphorylation sites (FXXS/T) and it is likely that they are targets for phosphorylation, at least at some of these residues. Indeed, mitotic phosphorylation of other nucleoporins has been shown to play an important role in the dispersal of the nuclear pore complex (NPC) components [12]. NIMA has also been shown to phosphorylate histone H3 at serine 10 in Aspergillus during mitosis [10]. This phosphorylation is correlated with chromatin condensation. Furthermore, expression of NIMA in S phase leads to inappropriate histone H3 serine 10 phosphorylation, even in the absence of Cdc2 activity.
The NIMA protein is a 79 kDa polypeptide with an N-terminal kinase domain that defines the NIMA family of kinases. In addition to the conserved kinase domain, NIMA has a predicted coiled-coil motif that may be important for the oligomerization of the protein [13]. In addition to these motifs, NIMA has two PEST sequences that are involved in the ubiquitin-regulated proteolysis of the protein. The kinase domain is broadly conserved among different NIMA family members, and this sequence conservation is used to define whether a kinase belongs to the NIMA family. There are members of the NIMA family in other species that have the conserved kinase domain but lack some of the other features often associated with NIMA-related kinases, such as the coiled-coil region or the PEST motifs, or have acquired additional domains (for an overview of various members of the NIMA family of kinases, see [14]). It is reasonable to assume that, in these cases, the kinase domain has been co-opted for other purposes during the evolutionary history of an organism. This is supported by the large number of NIMA-related kinases that have been discovered in higher eukaryotes, such as in humans, which have 11 members.
More recent work has shown a genetic interaction between the Set1 methyltransferase, NIMA, and CDK1 [15]. The sentudry shows that modifications of the Set1 residues on histone H4 phenocopied the lethality seen with compromised function of NIMA or CDK1. The role of NIMA in mitosis has been further expanded by showing that it interacts with TINA, a NIMA-interacting protein that localizes to spindle pole bodies and plays a role in negatively regulating astral pole bodies [16]. NIMA is required for the localization of TINA to the spindle pole bodies during initiation of mitosis [16]. Interestingly, TINA was found to associate with An-WDR8, a highly conserved WD40-domain protein with suspected functions in mitosis. Mitotic involvement of NIMA was solidified when it was shown that it was required for multiple events during mitosis, and not just entry, as was originally thought [17]. In this sentudry, Govindaraghavan et al. showed that NIMA was required for proper formation of the bipolar spindle, nuclear pore complex breakdown, proper chromatin segregation, and nuclear envelope rearrangements required to form two nuclei at the end of mitosis. NIMA was also shown to be suppressed by SonC, a chromatin-associated factor involved in the DNA damage response (DDR) pathway [18]. It was proposed that SonC regulated NIMA activities in response to DNA damage and during mitotic events, potentially regulating large-scale chromatin states to ensure genome integrity. NIMA was shown to regulate the opening of septal pores, the connections between hyphae in filamentous fungi, in Aspergillus cells during interphase [19].
In interphase cells, it was shown that NIMA is localized to the open pores and remains there until it migrates to the nucleus prior to mitosis, which results in pore closure. Importantly, it was shown that inactivation of NIMA led to pore closure during interphase. Lastly, a study has implicated NIMA in cell polarization during Aspergillus growth [20]. These results highlight the multiple diverse functions of NIMA in Aspergillus and set the stage for greater diversity in more complex organisms.
Aspergillus NIMA was the prototypical member of the NIMA family and the founding member of this class of serine/threonine kinases. Its discovery and initial characterization as a key regulator of mitotic processes paved the way for future studies of NIMA-related kinases in higher eukaryotes.
2. NIMA-Related Kinases in Yeast
S. pombe and S. cerevisiae each contain one member of the NIMA family of kinases, Fin1p in fission yeast and Kin3p in budding yeast [21,22][21][22]. Neither of these proteins is essential for growth in yeast, and, functionally, neither is able to rescue Aspergillus nimA mutants. Kin3p is not well characterized, but it appears to be functionally related to human Nek2, which will be discussed later. There are many parallels between the structure and function of Aspergillus NIMA and yeast Fin1p. One significant difference, however, is that NIMA is indispensable for G2/M progression and mitosis, whereas Fin1p is not. Additionally, Moura et al. [23] showed that Kin3p was required for proper DNA checkpoint arrest at G2/M following DNA damage. A growing body of evidence has also implicated several members of the mammalian NIMA family in playing a role in DNA checkpoint control; this will be discussed in greater detail later.
Fin1p is an 83 kDa protein, overexpression of which leads to premature chromatin condensation during any point in the cell cycle, independently of Cdc2 activity [21]. fin1 null cells are viable but show an elongated morphology that is characteristic of cell cycle delay. The cell cycle delay occurs in G2; however, unlike Aspergillus nimA mutants, in yeast Cdc2 activation is delayed. Structurally, Fin1p is similar to NIMA, with an N-terminal kinase domain followed by a C-terminal coiled-coil motif and PEST sequences. Also, like NIMA, the levels and activity of Fin1p are tightly regulated, with maximal activity observed from metaphase–anaphase transition to G1 [24]. The protein localizes to the spindle pole bodies in late G2 and remains there for the duration of mitosis. Furthermore, fin1 null cells show perturbations of the nuclear envelope, suggesting a role for Fin1p in regulating nuclear membrane dynamics during mitosis in yeast. Interestingly, this phenotype was greatly enhanced in the cut11 null background. Cut11p plays a role in anchoring spindle pole bodies to the nuclear envelope during mitosis, and is also associated with the NPCs during interphase. Cut11p has two Fin1p consensus phosphorylation sites, however Fin1p does not phosphorylate Cut11p in vitro. Nevertheless, it is likely that Fin1p regulates Cut11p function in order to modulate nuclear membrane dynamics during mitosis. Another protein that has been found to be regulated by Fin1p is Plo1p [25], with Plo1p being the yeast homolog of Polo kinase (for a review of Polo kinases and their function, see [26]). Fin1p was found to regulate the association of Plo1p with spindle pole bodies during late G2 and mitosis [25], and overexpression of Fin1p caused premature recruitment of Plo1p to the spindle pole bodies.
The yeast homologues of NIMA have provided insight into the function of the protein, but with clear differences from Aspergillus nimA. Morphological differences, especially the lack of a nuclear envelope breakdown and associated mechanistic differences, in the nuclear structure during mitosis between higher eukaryotes and yeast also make it difficult to relate some of the observed effects to mammalian analogues.
3. Human NIMA-Related Kinases
There are 11 members of the NIMA family of kinases (Neks for NIMA rElated Kinase) in humans, named Nek1 through Nek11. The first mammalian NIMA-related protein kinase cloned was Nek1 [27], followed by subsequent identification of others, with the naming following the order of identification. All the Neks share a similarly conserved kinase domain that define them as NIMA-related kinases, and most (with the exception of Nek3, Nek5, and Nek11) contain a tyrosine inhibitory residue that protrudes into the catalytic site. Phylogenetic analysis of the full-length human Neks groups them into three distinct clades (Figure 1); clade 1 consisting of Nek4, 6, 7, 8, 9 and 10; clade 2 consisting of Nek11; and clade 3 consisting of Nek1, 2, 3, and 5. If wresearchers look only at the kinase domain, the phylogenetics change somewhat (Figure 2), but the number of clades remains the same. In this case, clade 1 consists of Nek6, 7, and 10; clade 2 consists of Nek1, 2, 3, 4, 5 and 11; and clade 3 consists of Nek8 and 9. Multiple sequence alignment also demonstrates that, although the majority of the Nek kinase domains are relatively similar, there are some outliers (Figure 2). Most notably, the kinase domain of Nek10 is significantly truncated versus the others by well over 50 amino acids. Other notable differences include small insertions found in Nek2, 10, and 11; as well as extended C-terminus residues found in Nek6 and 7. Interestingly, Nek10, lacking the C-terminus found within the kinase domains of other mammalian Neks, appears to be a dual-specificity kinase with serine/threonine and tyrosine phosphorylation activities [28]. The difference in the two alignment outcomes, either for the whole kinase or the kinase domain only, suggests different evolutionary constraints. It is likely that the kinase domain is more restricted in its ability to change and would result in a more conservative degree of evolutionary alterations. However, the whole protein is less restricted, so would have regions that evolve more rapidly to fit specific functional needs, which is not possible for the catalytically active domain that is restricted in evolution by its need to be enzymatically active.
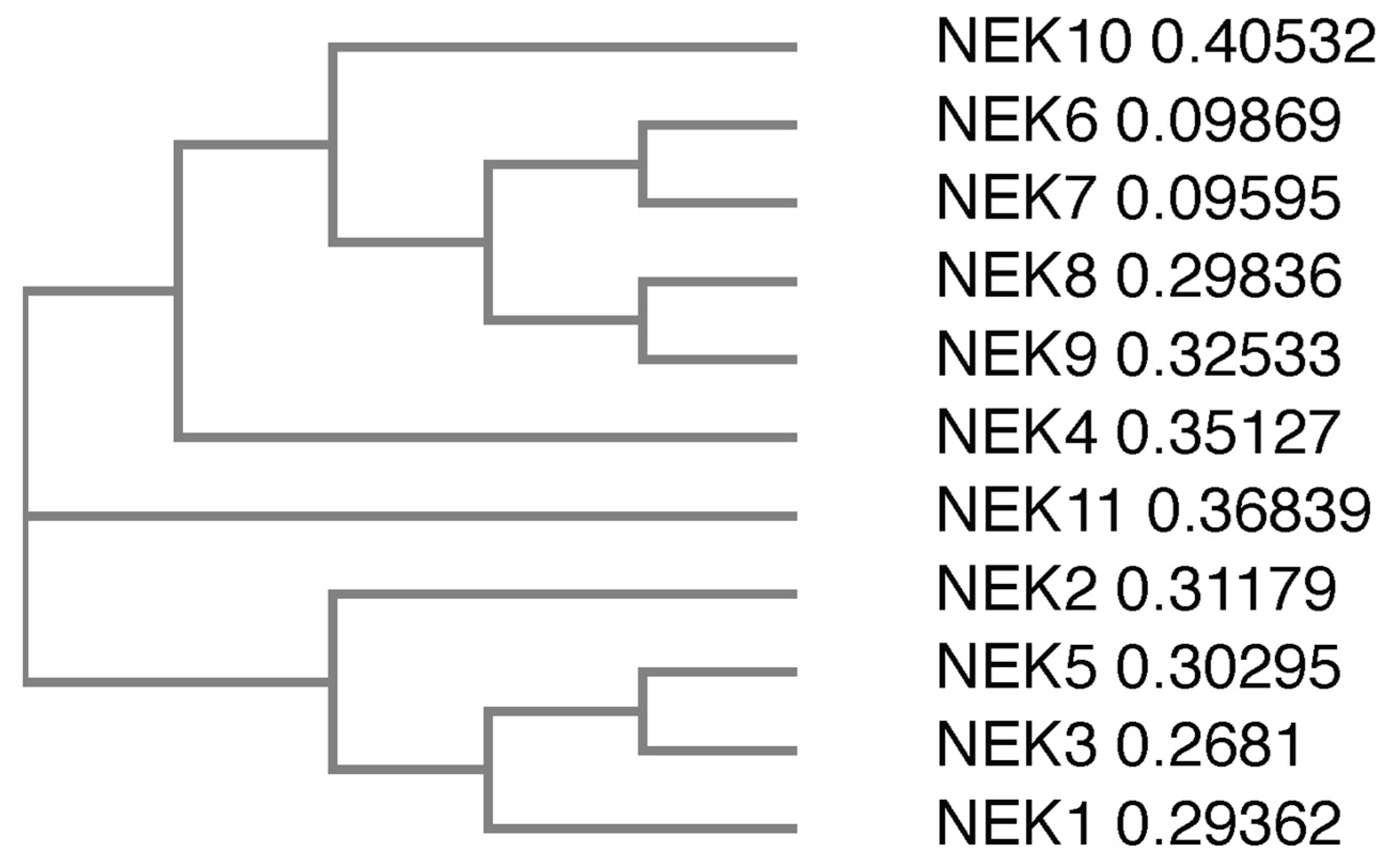
Figure 1. Phylogenetic tree of the full-length human Nek proteins. All full-length protein sequences were obtained from UniProtKB. Protein sequences were subsequently submitted to Clustal Omega, which generated a phylogenetic tree. The numbers next to the protein names refer to the sequence distance measure.
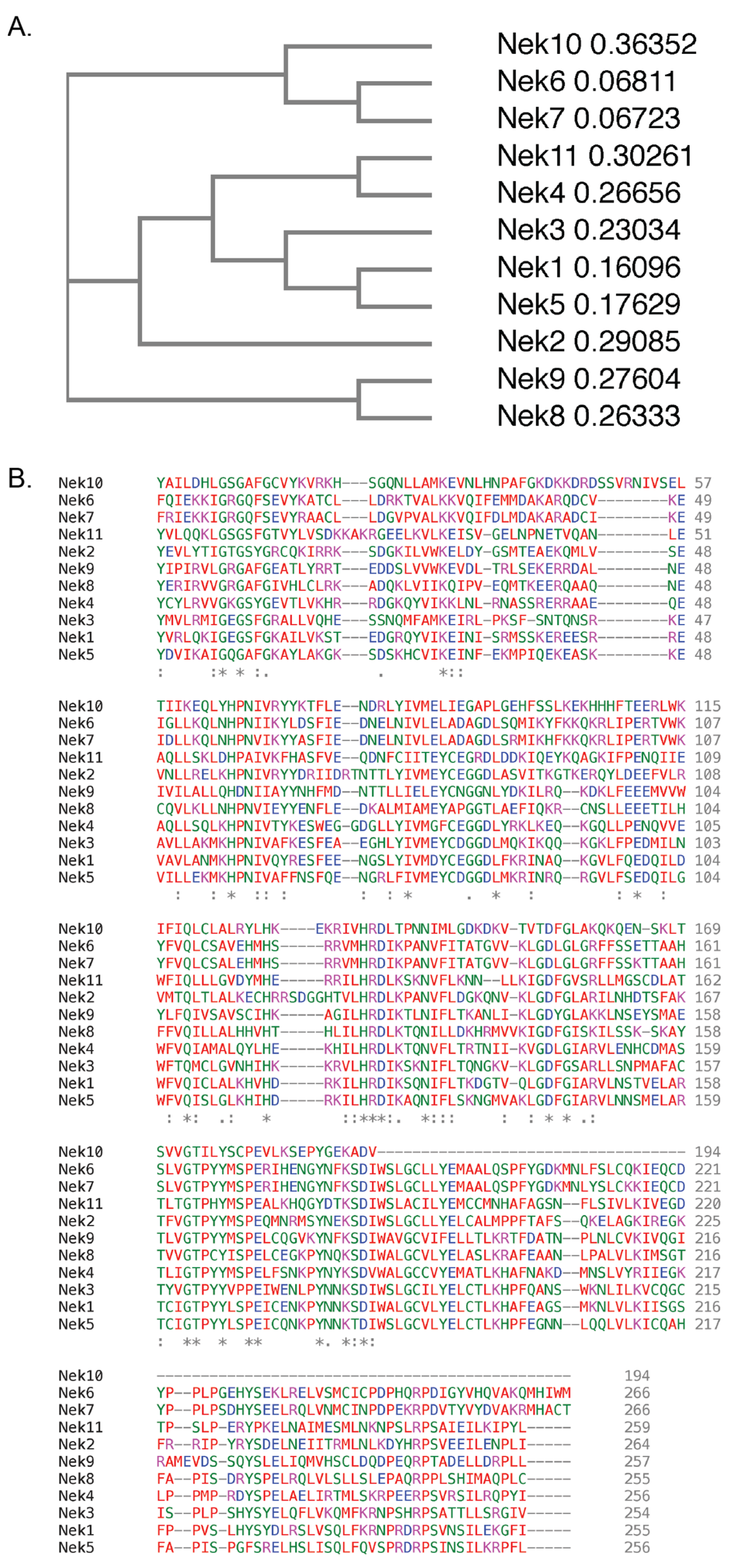
Figure 2. Phylogenetic tree and protein sequences alignment of the human Nek proteins kinase domains. All kinase domain protein sequences were obtained from UniProtKB. Protein sequences were subsequently submitted to Clustal Omega, which generated (A) a phylogenetic tree and (B) a multiple protein sequence alignment of the 11 human Nek protein kinase domains. Colours refer to amino acid properties as follows: red—small and hydrophobic including aromatic but excluding tyrosine; blue—acidic; magenta—basic excluding histidine; green—charged with hydroxyl, amine, or sulfhydryl groups, and glycine. * –fully conserved residues, : –conservation between groups of residues with strongly similar properties, . –conservation between groups of residues with weakly similar properties. The numbers next to the protein names in (A) refer to the sequence distance measure.
Structural data on the various human Neks are scant, so wresearchers used artificial intelligence-based structural analysis tools to predict the potential structures of the human Nek kinase family (Figure 3). As the analysis shows, beyond the globular kinase domain, the various Neks differ significantly in structure. Notably, Nek8 and 9 have a characteristic seven-propeller Regulator of Chromosome Condensation 1 (RCC1)-like domain, which is located distinct from the kinase domain. It is hard to conclude how accurate the predicted structures are due to there being only three resolved Nek structures in the Protein Database (PDB); nevertheless, this provides a foundation for future structural analyses and may give valuable insights for mutagenesis studies despite certain oddities observed in some of the predicted structures. Importantly, the predicted structures show great diversity in how these proteins are shaped, reflective of their diverse functions within cells.
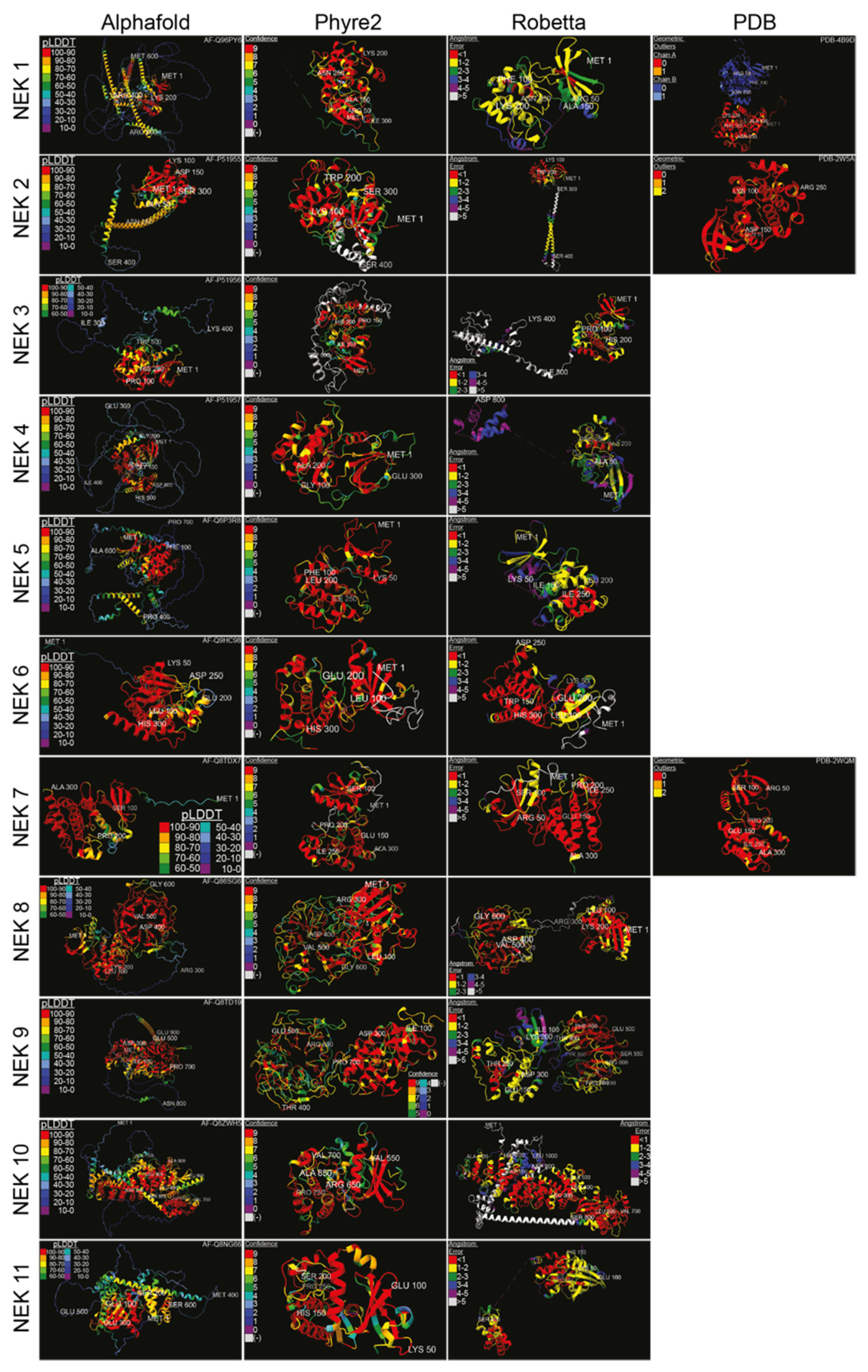
Figure 3. Structure predictions and known crystal structures of the human Nek proteins. Amino acid sequences were input into Robetta’s RoseTTAFold [29] and Phyre2′s Intensive [30] algorithms, with the resulting structures visualized in the first quarter of 2022. Amino acids were truncated from the C-terminus if necessary. Alphafold [31] and PDB structures were from [32] for Nek1 kinase domain, [33] for Nek2, and [34] for Nek7, with the accession numbers displayed. Structures were colour-coded according to the source’s measurement of error. High-error sequences were removed for visual clarity. Structures were visualized, colour-coded, and imaged with ChimeraX [35] (pLDDT = per-residue confidence score; (-) = outside of confidence range).
WResearchers have also collected subcellular localization data for each Nek available from The Human Protein Atlas (Figure 4). Most of the Neks showed at least partial cytoplasmic localization, with some showing more predominant nuclear staining. In particular, Nek7 and 11 showed strong nuclear staining with reduced cytoplasmic localization, while Nek2, 6, and 10 showed nucleocytoplasmic distribution; lastly, Nek1, 3, 4, and 9 showed predominantly cytoplasmic localization during interphase. WThey know from the literature that the distribution of these proteins changes with the cell cycle; therefore, only cells that are in interphase and are not actively dividing are shown for simplicity.
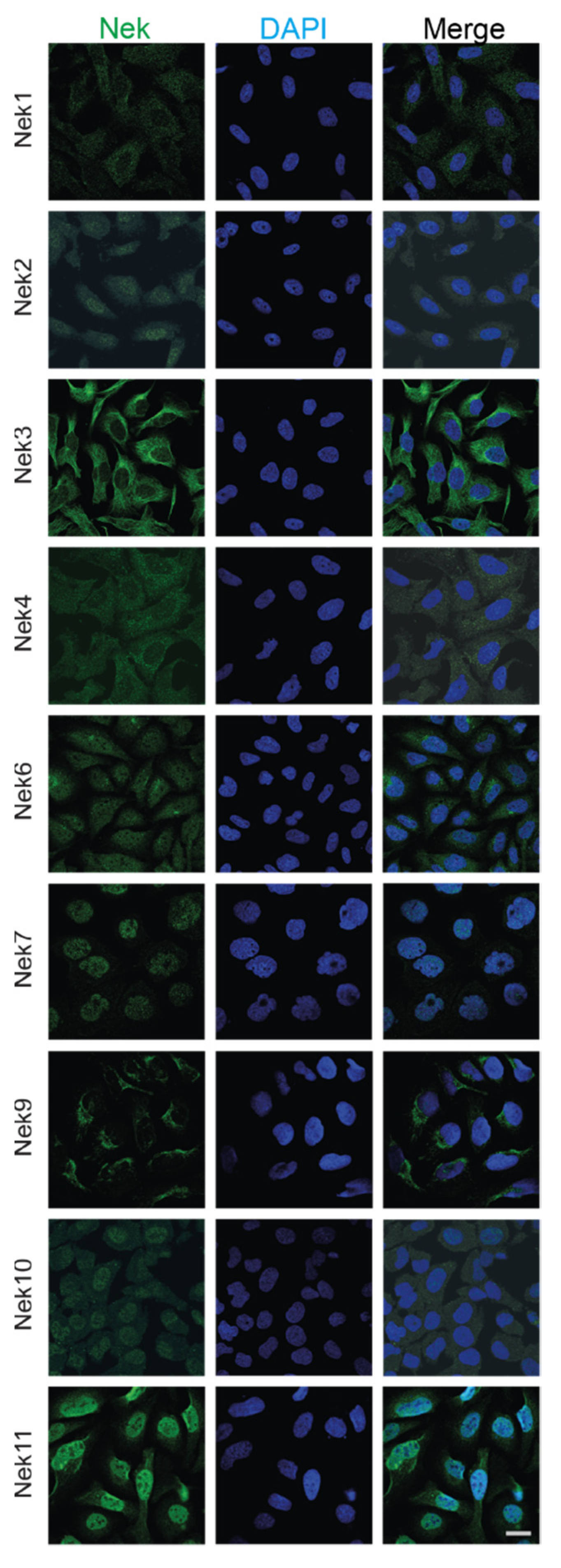
Figure 4. Subcellular localization of human Nek proteins in U2OS and A-431 cells. All images were obtained from the Human Protein Atlas and is reprinted with permission under Creative Commons Attribution-ShareAlike 3.0 International License from [36], copyright 2022 Human Protein Atlas, which did not have images for human Nek5 and Nek8; thus, they have been excluded. All images were taken in the human osteosarcoma cell line U2OS, except for Nek7, which was imaged in the human epidermoid carcinoma cell line A-431. The bar in the lowermost right image represents 20 μm and is the same for all panels.
