Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Conner Chen and Version 4 by Conner Chen.
When oxygenic photosynthesis evolved around 2.4 billion years ago, it is believed that all the oxygen-evolving organisms used the ribulose bisphosphate carboxylase-oxygenase (Rubisco)- photosynthetic carbon reduction cycle (PCRC) despite there being five other major CO2 assimilation pathways. Rubisco-PCRC was dominant, with the highest specific activity in presence of carbon dioxide at high concentrations without the presence of oxygen.
- Rubisco
- Synthetic Carbon Pathways
- Malonyl-CoA-Oxaloacetate-Glyoxylate (MOG)
- photosynthetic carbon reduction cycle
- thermodynamic bottlenecks
- thermodynamics
- Metabolic Regulation
- Metabolic Compatibility
- Energetic Efficiency
- carbon dioxide assimilation
1. Photosynthesis and Rubisco: An Evolutionary Perspective
When oxygenic photosynthesis evolved around 2.4 billion years ago, it is believed that all the oxygen-evolving organisms used the ribulose bisphosphate carboxylase-oxygenase (Rubisco)- photosynthetic carbon reduction cycle (PCRC) despite there being five other major CO2 assimilation pathways [1][2]. Rubisco-PCRC was dominant, with the highest specific activity in presence of carbon dioxide at high concentrations without the presence of oxygen. The other five pathways are (a) reverse tricarboxylic acid cycle (TCAC), (b) Hydroxypropionate, (c) Hydroxypropionate-4-hydroxybutyrate, (d) dicarboxylate-4-hydroxybutyrate, and (e) Wood–Ljungdahl pathway. What is unique about the Rubisco-PCRC pathway compared to the other five pathways is that oxygen is a competitive inhibitor for the Rubisco-PCRC pathway, while it is not for all the other five pathways. Rubisco enzyme is present both in prokaryotes and eukaryotes [3][4][5][6]. Four major forms of Rubisco have been identified (I, II, II/III, and IV) that are known to catalyze carboxylation and oxygenation of RuBP (Ribulose 1,5 Bisphosphate) [7][8]. Form II of Rubisco is associated with anoxygenic photosynthesis by proteobacteria before the evolution of oxygenic photosynthesis. The rise in oxygen concentration in the atmosphere with the proliferation of cyanobacteria over one billion years ago caused a decrease in CO2, ushering in the Great Oxidation Event (GOE) [1]. This change in atmospheric oxygen led to the evolution of oxygenic photosynthesis. The first time when Rubisco was exposed to oxygen led to the oxygenation of RuBP that in turn produced 2 phosphoglycolates, ushering in the evolution of the photorespiration pathway [1]. This photosynthesis with the evolution of oxygen eased the evolution of aerobic respiration, which is a bio-energetically more efficient process in comparison to anaerobic fermentation [1]. It is believed that the evolution of form I of Rubisco is closely linked to the increase in oxygen in the atmosphere [1]. The lowering of carbon-dioxide-to-oxygen ratios in the aerobic environment led to the evolution of more carbon-dioxide-specific enzymes starting to take place.
2. Can Engineering Photosynthesis Provide Solution?
Plants play a major role in fixing the global carbon footprint. Modern agriculture techniques with the use of fertilizer and irrigation provide evidence that carbon fixation could be a rate-limiting step. Many C3 plants when exposed to twice the carbon dioxide concentration have been shown to produce a significantly higher amount of biomass [9]. This suggests that plant growth could be regulated by manipulating biochemical pathways related to carbon fixation [10]. Studies have shown that Arabidopsis grew faster with more soluble sugars and enhanced shoot and root biomass when the natural photorespiration pathways were replaced with bacterial photorespiration pathways [11]. In another study with tobacco plants overexpressing an enzyme (sedoheptulose-1,7-bisphosphatase) that operates in the reductive pentose phosphate cycle, (rPP); showed enhanced biomass as well as a 30% increase in photosynthesis [12][13].
3. Why Is It Important to Look beyond Rubisco?
Diverse autotrophic organisms use the reductive pentose phosphate cycle (rPP cycle) for carbon dioxide assimilation. This cycle is rate-limited by the low catalytic rate of Rubisco (ribulose-1,5-bisphosphate carboxylase/oxygenase) [14][15][16]. The turnover of Rubisco and the low catalysis rate are negatively correlated, which suggests that the Rubisco enzyme is perhaps naturally optimized after evolving for millions of years [16][17][18]. Hence, optimizing Rubisco using genetic engineering may not prove to be a profitable endeavor [18]. This also suggests that it is imperative to look beyond the rPP cycle and identify Rubisco-independent pathways that have a higher carbon fixation rate.
4. What Are the Other Natural Carbon-Fixing Metabolic Pathways?
To date, apart from the rPP cycle, there are five other natural carbon-fixing metabolic pathways that have been identified. They are (a) 3-hydroxypropionate cycle; (b) reductive tricarboxylic acid (rTCA) cycle; (c) oxygen sensitive reductive acetyl-CoA (rAcCoA) pathways) (Figure 12); (d) dicarboxylate/4-hydroxybutyrate cycle (Figure 23); and (e) 3-hydroxypropionate/4-hydroxybutyrate cycle (Figure 23) [16][18][19][20].
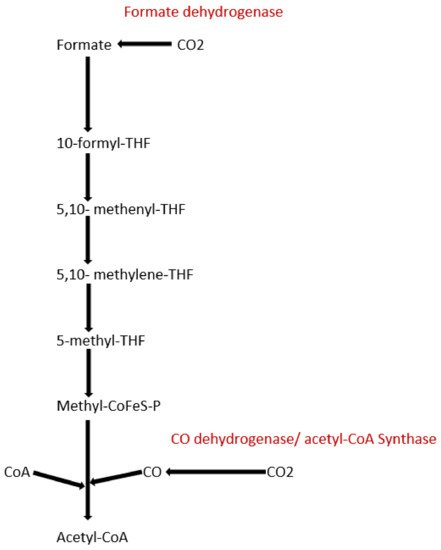
Figure 12. Schematic representation of reductive acetyl-CoA pathway.
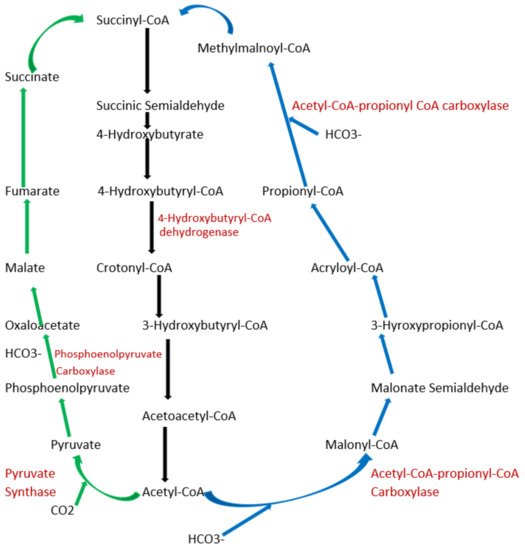
Figure 23. Schematic representation of dicarboxylate/4-hydroxybutyrate and 3-hydroxypropionate/4-hydroxybutyrate cycle. Green arrows indicate the dicarboxylate/4-hydroxybutyrate metabolic pathway, and blue arrows represent 3-hydroxypropionate/4-hydroxybutyrate cycle pathway. Black arrows represent common metabolic pathway between the two pathways.
5. Exploring Other Alternatives: Introduction to Synthetic Pathways
To explore other alternatives, it is important to be aware of various factors that are to be considered while searching for alternative pathways. Studies have been conducted exploring approximately 5000 enzymes from the KEGG database to seek potential carbon fixation pathways [21]. The pathways were analyzed and evaluated based on four criteria, i.e., kinetics, thermodynamics, the efficiency of energetics, and topological compatibility. All the criteria were compared to the six naturally occurring carbon fixation pathways to validate the results [22]. Other studies have included a few other factors in addition to the ones mentioned above such as toxicity of metabolites produced and regulation of the cycle [23][24][25].
Combining the six criteria from the above studies would provide a comprehensive matrix to analyze and establish novel carbon fixation pathways.
5.1. How to Use the Criteria Matrix to Select Synthetic Carbon Pathways?
5.1.1. Kinetic Analysis
While considering the kinetics of theoretical pathways, the Kcat/Km of each of the enzymes should have a high value for Kcat and a low value for Km. This will ensure that there is high enzyme specificity in addition to a high rate of catalysis [26]. Now the question is, where to start? The best way to get started is to look for rate-limiting steps and identify enzymes associated with them. An example could be finding carboxylation enzymes that have higher specific activity and have higher affinities for carbon dioxide and bicarbonate ions under ambient conditions. Enzymes such as phosphoenolpyruvate (PEP) carboxylase, pyruvate carboxylase, acetyl-CoA, and propionyl-CoA carboxylases are some of the examples.
5.1.2. Resource Consumption and Energetic Efficiency
Too much dependence on kinetics could be detrimental, as it does not provide information on resource consumption for each pathway. It is important to calculate the energetic cost simultaneously for each pathway under consideration. The energetic cost for each pathway can be classified into NADPH cost and ATP cost. While calculating NADPH cost, one needs to consider the number of moles of NADPH consumed to produce one mole of a product. Under NADPH, other redox carriers such as ferredoxins and FADH2 are also counted. Similarly, for the ATP cost, consumption of 1 mole of non-redox carriers such as nucleotide triphosphates (NTPs), coenzyme A thioesters, and phosphoesters for the production of 1 mole of product needs to be calculated.
5.1.3. Thermodynamic Analysis
While analyzing kinetics and resource consumption, it is imperative to concurrently conduct a thermodynamic analysis of the pathways. At ambient conditions and carbon dioxide concentrations, the Gibbs free energy should be negative, i.e., (∆Go << 0). Some of the natural pathways such as the rTCA cycle [27] and the rAcCoA pathway do not function under ambient carbon dioxide concentrations if the ranges of pH and ionic strengths are broad, as the (∆Go > 0). Interestingly, the rTCA cycle can function under a broad range of pH and ionic strengths if the carbon dioxide concentrations are 100 times higher than the ambient concentration. Similarly, the rAcCoA pathway can function at pH < 8 under high carbon dioxide concentrations. This is possible especially for C4 and CAM plants that use mechanisms to concentrate carbon dioxide. Hence, it could be safely concluded that both the rTCA cycle and the rAcCoA pathway work under anaerobic conditions. This is important, as it is a known fact that anaerobes use pathways that have low energetic yields. Hence, using such pathways would limit the success in enhancing carbon fixation yield. The only way these two pathways can be feasible is if the plants are grown under high-carbon-dioxide conditions.
5.1.4. Analysis of Distributed Thermodynamic Bottlenecks
Theoretically, analyzing the energetics of the whole pathway as stated above can predict whether the pathway is feasible or not. It can also help tous predict when there are no constraints on the concentration of various metabolites. In reality, the concentrations of metabolites have an upper and lower limit. Hence, analysis of the energetics of the overall pathway does not guarantee feasibility. There is always a chance the predictions can fail despite being ∆Go < 0, as there are sub-pathways within the major pathway that are concentration-dependent. These sub-pathways are known as thermodynamic bottlenecks.
To analyze thermodynamic bottlenecks, the major pathway is analyzed by analyzing the Gibbs free energy change at an ambient temperature of the sub-cycles. ∆Go of each sub-cycle is experimentally analyzed at multiple pH ranges and multiple ionic concentrations without accounting for the ATP hydrolysis. If the ∆Go of all the sub-pathways for all the pH and ionic range is negative, then it can be safely concluded that the following major pathway is feasible and a promising candidate for future carbon fixation studies [28][29][30].
5.1.5. Metabolic Regulation and Compatibility
As the pathways are analyzed, it is important to simultaneously consider the integration and compatibility of the network with other pathways inside the cell. The key parameters to consider are a number of enzymes involved in the fixation cycle and conducting random sampling followed by flux analysis to determine compatibility with the other endogenous network in the cell. When attempting to design a synthetic pathway, it is lucrative to engineer it in such a way that there are as few of these manual checkpoints as possible to reduce the number of complications that would arise from the need to maintain them. However, there are exceptions. An instance where having regulation points would be useful is when one wants to limit the number of irreversible enzymes that produce intermediates to prevent flux imbalances [27].
5.1.6. Analysis of Metabolites
The metabolic intermediates should be as unreactive and non-toxic to the cell as possible. Not only this but having the intermediates be considerably hydrophobic should be avoided in order to prevent them from leaking out from within the cell [5]. Additionally, metabolites containing low concentrations of cofactors should be used sparingly, if at all [27]. This will reduce the number of challenges that will arise from having to regulate cofactors and prevent them from depleting.
5.1.7. Application of Synthetic Pathways
Malonyl-CoA-Oxaloacetate-Glyoxylate (MOG) is one of the synthetic pathways discovered using the method mentioned above. Promising candidates for carbon-fixing enzymes were identified, and the pathway was predicted to be thermodynamically feasible [29][30]. MOG showed similarity with carboxylation products, namely Malonyl-CoA and oxaloacetate. It was called the Malonyl-CoA-Oxaloacetate-Glyoxylate (MOG) pathway because of their common carboxylation and export products. There are limitations like some other pathways, such as requiring a certain temperature for function, and glyoxylate produces carbon dioxide via decarboxylation [27][5].
