Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 4 by Olga V Anatskaya and Version 3 by Conner Chen.
Polyploidy or whole-genome duplication (WGD) is widespread in nature, agriculture and aquaculture, normal physiology, regeneration, aging, and pathology. WGD results from the premature termination of the cell cycle or cell fusion. If WGD occurs in germ cells, the progeny organisms become completely polyploid, if in somatic cells, the somatic polyploidy arises in certain tissues of a given organism. Polyploidization leads to long-term consequences both in evolution (organismal) and ontogenesis (somatic). In evolution, WGD is one of the main sources for the growth of organismal complexity and evolutionary plasticity
- evolutionary conserved features
- complexity
- epigenetic changes
- biological plasticity
- adaptation
- stress resistance
- regeneration
- carcinogenesis
- aging
- developmental programming
- cardiovascular disease
1. Introduction
Polyploidy or whole-genome duplication (WGD) is widespread in nature, agriculture and aquaculture, normal physiology, regeneration, aging, and pathology [1][2][3]. WGD results from the premature termination of the cell cycle or cell fusion [2][4]. If WGD occurs in germ cells, the progeny organisms become completely polyploid, if in somatic cells, the somatic polyploidy arises in certain tissues of a given organism. Polyploidization leads to long-term consequences both in evolution (organismal) and ontogenesis (somatic) [1].
In evolution, WGD is one of the main sources for the growth of organismal complexity and evolutionary plasticity [5][6]. In ontogenesis, genome duplication can result from a trade-off between cell proliferation, stress, and tissue-specific functional load [7][8][9][10][11]. These trade-offs are supposed to be a connecting link between the role of genome duplications in normal and pathological conditions. They present a special case of the main contradiction of multicellularity (MCM) existing between the cellular and organismal levels [12]. The cellular level is presented by cell proliferation, whereas the organismal level is embodied in the control of cell proliferation and the tissue-specific functional load. The similarity between the organismal and somatic polyploidy is the increase in stress resistance, plasticity, induction of pathways of morphogenesis and longevity, epigenetic and metabolic changes.
2. Evolution
2.1. General Picture
One of nature’s greatest mysteries is how simple bacteria could give rise to such complex creatures as humans. The fundamental law of evolution (natural selection of undirected genetic changes) follows from the ‘vertical’ copying of genetic information (with small random changes). The vertical copying increases the organism’s numbers, thus resulting in competition for resources and natural selection. However, the growth of complexity does not follow from the vertical copying of information. It depends on the second fundamental copying type, which is orthogonal to the first.
The increase in the information amount can occur due to duplication not only of the whole genome. Susumu Ohno was the first to attract attention to the role of WGD in evolution [13]. In honor of Ohno, the genes arising from the organismal polyploidization are called ‘ohnologs’ [14]. His main idea was that as a result of duplication, an additional gene copy is freed from purifying selection thereby acquiring evolutionary plasticity. He paid special attention to WGD as it completely preserves gene regulatory regions and an initial gene dosage balance, thereby providing a wide foundation for systemic evolutionary experiments. In addition, polyploidization plays a special role in the increase in complexity due to the certain properties of cellular network growth in the case of WGD as compared with the single-gene duplication [12], which will be discussed below.
Now it is firmly established that there were two rounds of ancient WGD in the vascular plant lineage (390 and 300 Mya) and in the vertebrate lineage between chordates and vertebrates (both about 500 Mya, near Cambrian explosion) [14]. In teleost fishes, there was a third WGD (300 Mya), while in salmonid fishes, a fourth (95 Mya) [15][16]. Furthermore, the more recently formed polyploid strains, species, and above-species lineages are widespread in nature, agriculture, and aquaculture. They are frequent in plants, prokaryotes, protists, fungi, invertebrates, and ectotherm vertebrates [5][17][18]. In vertebrates, the record should be placed in the Xenopus genus where 26 of 27 species are polyploids, and in sturgeons where dodecaploid species with 380 chromosomes were found [19][20]. In reptiles, there are only triploid species (and they are very rare), which propagate by parthenogenesis [21]. In birds and mammals, polyploids can also arise after conception, yet die in early development [22]. One rodent species is sometimes mentioned as polyploid, however, this attribution is probably incorrect [23]. Notably, certain model organisms are lineage-specific polyploids (yeast, zebrafish, xenopus, Arabidopsis).
2.2. Cellular and Molecular Aspects
One of the main problems, which new polyploids encounter, is the disturbance of chromosome pairing in meiosis. This problem is alleviated in the case of allopolyploids where polyploidization occurs due to the hybridization of two related species. In allopolyploids, the chromosomes of parental species differ and therefore conjugate separately. Probably as a result of this feature, allopolyploids are more frequently established, albeit more rarely, compared to autopolyploids [18][24]. Polyploidization is followed by rediploidization, i.e., the process of converting a polyploid chromosome set into a diploid one. Rediploidization does not mean the reversal to the original diploid state but only the re-establishment of regular chromosome pairing due to chromosome rearrangement and DNA divergence [16][20]. This process is accompanied by gene loss (due to deletion or pseudogenization), subfunctionalization, and neofunctionalization [25][26]. Subfunctionalization is a partitioning of an initial gene function (e.g., making it tissue-/development stage-/condition-specific), whereas neofunctionalization is an acquisition of a novel function. Subfunctionalization and neofunctionalization lead to gene diversification, which should be accompanied by an integration of the work of a growing number of diversifying genes, i.e., an increase in regulatory complexity [27].
In genome-wide studies, subfunctionalization can be detected by the changes in gene expression and the properties of the interactome. For example, in the yeast interactome, the proteins encoded by ohnologs have a lower number of protein interactions compared to the single-gene duplicates and genes without duplicates (i.e., genes that lost the second ohnolog) [28]. The loss of interactions was proportional to their initial number and independent of the ohnolog position in the protein interaction network. The functional analysis of the overlapping and non-overlapping interactants in each pair of ohnologs revealed a sharp asymmetry in the regulatory functions. The regulatory functions occur more frequently in the non-overlapping interactants. These facts suggest that subfunctionalization was a prevailing trend in the evolution of retained ohnologs, and was accompanied by regulatory specialization.
The increase in regulatory complexity and elaboration of signal transduction systems is a hallmark of post-WGD evolution [29]. The predominant retention of regulatory and developmental genes subsequent to WGD can take place as relaxed selection allowed duplicated networks to be rewired, and to evolve novel functionality, thereby increasing biological complexity [5]. In the human interactome, the more ancient genes have a higher local and global centrality compared with newer genes, which indicates a gradual core-to-periphery evolutionary growth of the interactome [12]. The local centrality is the number of direct interactions of a protein (degree), while the global centrality is estimated by the number of protein-interaction paths going through a protein in the whole network (betweenness) or by the average path length to other proteins (closeness). However, there is a remarkable exception: the novel genes did not decline in the network centrality during the expansion of the multicellular organization from Bilateria to Euteleostomi (bony vertebrates) [12]. This plateau of interactome centrality is related to the series of whole-genome duplication. It indicates that in contrast to single-gene duplicates, which provide a gradual core-to-periphery network growth, in the case of WGD, the network core (that is richest in interactions) is also duplicated. Thus, the interactome complexity becomes higher in the case of growth by means of WGD, as compared with the growth by single-gene duplicates. The great diversity of vertebrates, which appeared after the two WGD rounds followed by sub- and neofunctionalization of ohnologs, can be considered as an extension of Cambrian explosion in the vertebrate lineage (reflected in the plateau of interactome centrality).
The human ohnologs are most strongly enriched in the bivalent genes and the genes related to development, neuronal membrane, and chromatin (Figure 1 and Figure 2). The bivalent genes, which have both activating and repressive epigenetic marks in their promoters and can switch on/off quickly, are key regulators of cellular networks [30][31]. These facts emphasize the important role of ohnologs in regulation.
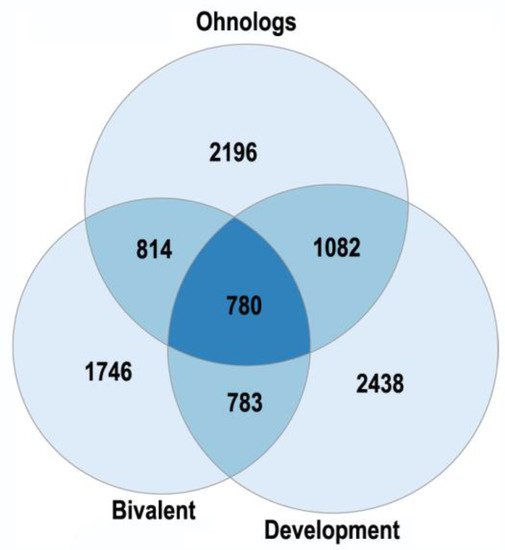
Figure 1. The enrichment of human ohnologs in bivalent genes and genes involved in multicellular organism development GO:0007275. Bivalent: p < 10−151, development: p < 10−67.
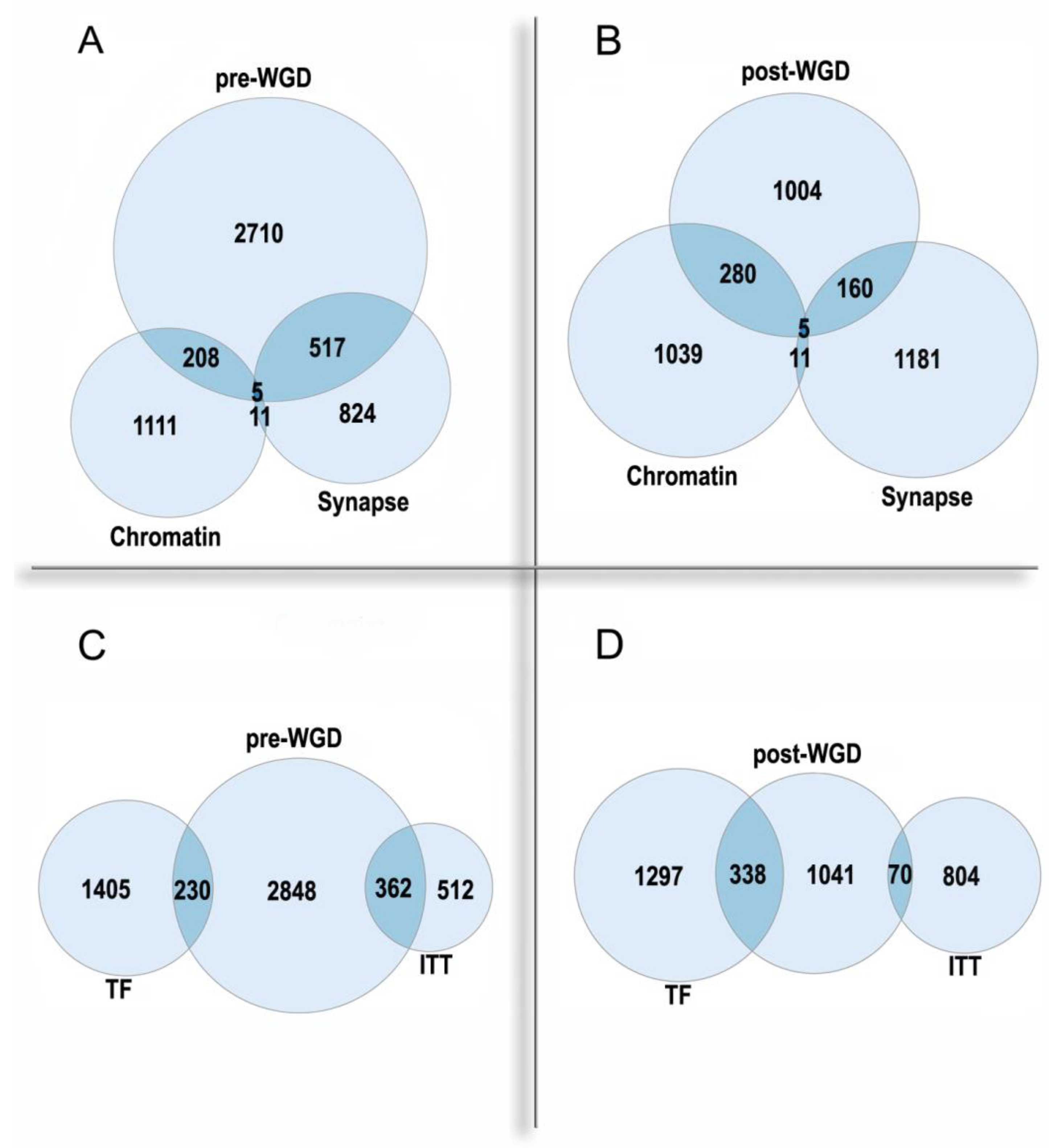
Figure 2. The enrichment of pre-WGD and post-WGD human ohnologs in functional gene groups. (A,B)—nuclear chromatin GO:0000790 and synapse GO:0045202. Chromatin: underrepresentation p < 0.01 in pre-WGD, enrichment p < 10−61 in post-WGD. Synapse: enrichment p < 10−71 in pre-WGD; enrichment p < 10−8 in post-WGD. (C,D)—transcription factors (TF) and ion transmembrane transporter activity GO:0015075 (ITT). TF: underrepresentation p < 10−7 in pre-WGD, enrichment p < 10−74 in post-WGD. ITT: enrichment p < 10−56 in pre-WGD, not significant p > 0.4 in post-WGD. Albeit both pre-WGD and post-WGD ohnologs are enriched in the synapse genes, there are significant differences in binomial proportions between pre-WGD and post-WGD ohnologs (p < 10−15).
The ohnologs present a unique model for the study of evolutionary speed in different functional gene groups (as WGD presents the common starting point). Albeit all ohnologs are of pre-WGD origin by definition, it is possible to distinguish those of them, which experienced an accelerated sub/neofunctionalization after the WGD, using the method of shallow phylostratigraphy (gene dating) [33]. These genes rapidly accumulated changes and can be mapped only to the post-WGD phylostrata by shallow phylostratigraphy. In other words, as a result of the rapid evolution, they lost strict orthology with the genes from phylogenetic lines branching before the WGD and retained it only with the genes from the post-WGD branches (they were called ‘post-WGD’ ohnologs).
Importantly, the synapse and the chromatin are the most enriched Cellular Component Gene Ontology categories in the pre-WGD and post-WGD ohnologs, respectively. Thus, ohnologs are involved in both regulatory levels of the organism: the nucleome and the connectome. The pre-WGD ohnologs show enrichment in the synapse genes and underrepresentation in the chromatin genes, whereas the post-WGD ohnologs show a stronger enrichment in the chromatin genes than in the synapse genes, with significant difference in these enrichments (Figure 2A,B). The most enriched GO Molecular Function in the pre-WGD ohnologs is the ion transmembrane transporter activity, whereas in the post-WGD ohnologs it is the transcription factors (Figure 2C,D). Thus, nuclear regulome shows a faster evolution than the molecular basis of neuronal signal transmission. Possibly, this is due to the main regulatory task in the nervous system being shifted from the molecular to the cellular level, and this shift can restrict the evolution of involved genes.
In addition to the increase in complexity, polyploids show enhanced evolutionary plasticity [35]. Large-scale species radiation in a given taxonomic group typically follows polyploidization events, indicating the potential of polyploidization for speciation [36]. However, whatever evolutionary perspectives of polyploids may be, natural selection is not teleological and works only for immediate adaptation. Polyploids show a higher resistance to environmental stress and diseases [37][38]. They are especially advantageous under dramatically challenging environments [39]. Thus, many ancient polyploidization events were concentrated near crucial historical periods, such as the Cretaceous–Paleogene boundary, when an asteroid hit the Earth resulting in the drastic alteration of climate and mass extinction [36].
2.3. Polyploidy in Agriculture and Aquaculture Biotechnology
Polyploidy was a key factor in the domestication of crops [26][40][41]. Domesticated plants have gone through more polyploidization events than their wild relatives [40]. Genetic plasticity of the polyploid genome and multicopy genes present a special advantage for domestication [36]. Among lineage-specific polyploids are such major crops as wheat, oat, maize, cotton, potato, legumes, banana, sugarcane, oilseed rape, strawberry, coffee, mustard, tobacco. The domesticated organisms are mostly allopolyploids (i.e., the results of polyploidization associated with hybridization). Hybridization is frequently accompanied by enhanced heterozygosity and hybrid vigor, while polyploidization restores the fertility of newly formed hybrids and contributes to the stabilization of the hybrid genome, fixing both heterozygosity and new hybrid characters [41].
Allopolyploidization presents a way for new organisms’ formation. Synthetic polyploids have been employed to increase beneficial traits, such as higher fitness, disease resistance, faster growth, and larger production compared to their natural diploids [42][43]. Thus, the synthetic hexaploid wheat presents a novel source of genetic diversity for multiple biotic stress resistance [38]. Many artificial polyploids are utilized commercially in aquaculture and most of them were created from natural polyploid fishes and shellfishes, especially cyprinids and salmonids [44]. There is a competition between parental genomes in polyploid hybrids, called ‘subgenome dominance’, when the less expressed subgenome tends to delete more genes and cis-acting elements compared to the counterpart subgenome [26][41]. In extreme cases, the parental genomes can even segregate separately in germ cell lineage and a hybrid state arises de novo in each generation [45][46]. The recursive production of de novo polyploid hybrids is used when allopolyploids are sterile or lose their hybrid vigor through inbreeding depression. For instance, an allohexaploid carp is massively propagated by crossing parental species in each generation [44].
References
- Gjelsvik, K.J.; Besen-McNally, R.; Losick, V.P. Solving the Polyploid Mystery in Health and Disease. Trends Genet. 2019, 35, 6–14.
- Donne, R.; Saroul-Aïnama, M.; Cordier, P.; Celton-Morizur, S.; Desdouets, C. Polyploidy in Liver Development, Homeostasis and Disease. Nat. Rev. Gastroenterol. Hepatol. 2020, 17, 391–405.
- Fox, D.T.; Soltis, D.E.; Soltis, P.S.; Ashman, T.-L.; Van de Peer, Y. Polyploidy: A Biological Force From Cells to Ecosystems. Trends Cell Biol. 2020, 30, 688–694.
- Dörnen, J.; Sieler, M.; Weiler, J.; Keil, S.; Dittmar, T. Cell Fusion-Mediated Tissue Regeneration as an Inducer of Polyploidy and Aneuploidy. Int. J. Mol. Sci. 2020, 21, 1811.
- Van de Peer, Y.; Mizrachi, E.; Marchal, K. The Evolutionary Significance of Polyploidy. Nat. Rev. Genet. 2017, 18, 411–424.
- Van de Peer, Y.; Ashman, T.-L.; Soltis, P.S.; Soltis, D.E. Polyploidy: An Evolutionary and Ecological Force in Stressful Times. Plant Cell 2021, 33, 11–26.
- Anatskaya, O.V.; Vinogradov, A.E. Genome Multiplication as Adaptation to Tissue Survival: Evidence from Gene Expression in Mammalian Heart and Liver. Genomics 2007, 89, 70–80.
- Anatskaya, O.V.; Vinogradov, A.E. Somatic Polyploidy Promotes Cell Function under Stress and Energy Depletion: Evidence from Tissue-Specific Mammal Transcriptome. Funct. Integr. Genom. 2010, 10, 433–446.
- Anatskaya, O.V.; Sidorenko, N.V.; Beyer, T.V.; Vinogradov, A.E. Neonatal Cardiomyocyte Ploidy Reveals Critical Windows of Heart Development. Int. J. Cardiol. 2010, 141, 81–91.
- Anatskaya, O.V.; Erenpreisa, J.A.; Salmina, K.A.; Vazquez-Martin, A.; Huna, A.; Nikolsky, N.N.; Vinogradov, A.E. Polyploidy Activates Biological Pathways Related to Morphogenesis in Mammalian Tissues. MOJ Immunol. 2018, 6, 90–93.
- Vazquez-Martin, A.; Anatskaya, O.V.; Giuliani, A.; Erenpreisa, J.; Huang, S.; Salmina, K.; Inashkina, I.; Huna, A.; Nikolsky, N.N.; Vinogradov, A.E. Somatic Polyploidy Is Associated with the Upregulation of C-MYC Interacting Genes and EMT-like Signature. Oncotarget 2016, 7, 75235–75260.
- Vinogradov, A.E.; Anatskaya, O.V. Evolutionary Framework of the Human Interactome: Unicellular and Multicellular Giant Clusters. Biosystems 2019, 181, 82–87.
- Ohno, S. Evolution by Gene Duplication; Springer: Berlin/Heidelberg, Germany, 1970; ISBN 978-3-642-86661-6.
- Singh, P.P.; Isambert, H. OHNOLOGS v2: A Comprehensive Resource for the Genes Retained from Whole Genome Duplication in Vertebrates. Nucleic Acids Res. 2020, 48, D724–D730.
- Inoue, J.; Sato, Y.; Sinclair, R.; Tsukamoto, K.; Nishida, M. Rapid Genome Reshaping by Multiple-Gene Loss after Whole-Genome Duplication in Teleost Fish Suggested by Mathematical Modeling. Proc. Natl. Acad. Sci. USA 2015, 112, 14918–14923.
- Robertson, F.M.; Gundappa, M.K.; Grammes, F.; Hvidsten, T.R.; Redmond, A.K.; Lien, S.; Martin, S.A.M.; Holland, P.W.H.; Sandve, S.R.; Macqueen, D.J. Lineage-Specific Rediploidization Is a Mechanism to Explain Time-Lags between Genome Duplication and Evolutionary Diversification. Genome Biol. 2017, 18, 111.
- Soppa, J. Polyploidy and Community Structure. Nat. Microbiol. 2017, 2, 16261.
- Hu, G.; Wendel, J.F. Cis-Trans Controls and Regulatory Novelty Accompanying Allopolyploidization. New Phytol. 2019, 221, 1691–1700.
- Evans, B.J.; Carter, T.F.; Greenbaum, E.; Gvoždík, V.; Kelley, D.B.; McLaughlin, P.J.; Pauwels, O.S.G.; Portik, D.M.; Stanley, E.L.; Tinsley, R.C.; et al. Genetics, Morphology, Advertisement Calls, and Historical Records Distinguish Six New Polyploid Species of African Clawed Frog (Xenopus, Pipidae) from West and Central Africa. PLoS ONE 2015, 10, e0142823.
- Du, K.; Stöck, M.; Kneitz, S.; Klopp, C.; Woltering, J.M.; Adolfi, M.C.; Feron, R.; Prokopov, D.; Makunin, A.; Kichigin, I.; et al. The Sterlet Sturgeon Genome Sequence and the Mechanisms of Segmental Rediploidization. Nat. Ecol. Evol. 2020, 4, 841–852.
- Moritz, C.; Bi, K. Spontaneous Speciation by Ploidy Elevation: Laboratory Synthesis of a New Clonal Vertebrate. Proc. Natl. Acad. Sci. USA 2011, 108, 9733–9734.
- Otto, S.P. The Evolutionary Consequences of Polyploidy. Cell 2007, 131, 452–462.
- Evans, B.J.; Upham, N.S.; Golding, G.B.; Ojeda, R.A.; Ojeda, A.A. Evolution of the Largest Mammalian Genome. Genome Biol. Evol. 2017, 9, 1711–1724.
- Barker, M.S.; Arrigo, N.; Baniaga, A.E.; Li, Z.; Levin, D.A. On the Relative Abundance of Autopolyploids and Allopolyploids. New Phytol. 2016, 210, 391–398.
- Venkatachalam, A.B.; Parmar, M.B.; Wright, J.M. Evolution of the Duplicated Intracellular Lipid-Binding Protein Genes of Teleost Fishes. Mol. Genet. Genom. MGG 2017, 292, 699–727.
- Cheng, F.; Wu, J.; Cai, X.; Liang, J.; Freeling, M.; Wang, X. Gene Retention, Fractionation and Subgenome Differences in Polyploid Plants. Nat. Plants 2018, 4, 258–268.
- Vinogradov, A.E. Global versus Local Centrality in Evolution of Yeast Protein Network. J. Mol. Evol. 2009, 68, 192–196.
- Vinogradov, A.E.; Anatskaya, O.V. Loss of Protein Interactions and Regulatory Divergence in Yeast Whole-Genome Duplicates. Genomics 2009, 93, 534–542.
- Corrochano, L.M.; Kuo, A.; Marcet-Houben, M.; Polaino, S.; Salamov, A.; Villalobos-Escobedo, J.M.; Grimwood, J.; Álvarez, M.I.; Avalos, J.; Bauer, D.; et al. Expansion of Signal Transduction Pathways in Fungi by Extensive Genome Duplication. Curr. Biol. 2016, 26, 1577–1584.
- Court, F.; Arnaud, P. An Annotated List of Bivalent Chromatin Regions in Human ES Cells: A New Tool for Cancer Epigenetic Research. Oncotarget 2017, 8, 4110–4124.
- Jeon, A.-J.; Tucker-Kellogg, G. Bivalent Genes That Undergo Transcriptional Switching Identify Networks of Key Regulators of Embryonic Stem Cell Differentiation. BMC 2020, 21, 614.
- Vinogradov, A.E. “Genome Design” Model and Multicellular Complexity: Golden Middle. Nucleic Acids Res. 2006, 34, 5906–5914.
- Vinogradov, A.E.; Anatskaya, O.V. Growth of Biological Complexity from Prokaryotes to Hominids Reflected in the Human Genome. Int. J. Mol. Sci. 2021, 22, 11640.
- Hu, H.; Miao, Y.-R.; Jia, L.-H.; Yu, Q.-Y.; Zhang, Q.; Guo, A.-Y. AnimalTFDB 3.0: A Comprehensive Resource for Annotation and Prediction of Animal Transcription Factors. Nucleic Acids Res. 2019, 47, D33–D38.
- Mattenberger, F.; Sabater-Muñoz, B.; Toft, C.; Fares, M.A. The Phenotypic Plasticity of Duplicated Genes in Saccharomyces Cerevisiae and the Origin of Adaptations. G3: Genes Genomes Genet. 2017, 7, 63–75.
- Zhang, K.; Wang, X.; Cheng, F. Plant Polyploidy: Origin, Evolution, and Its Influence on Crop Domestication. Hortic. Plant J. 2019, 5, 231–239.
- Ruiz, M.; Quiñones, A.; Martínez-Cuenca, M.R.; Aleza, P.; Morillon, R.; Navarro, L.; Primo-Millo, E.; Martínez-Alcántara, B. Tetraploidy Enhances the Ability to Exclude Chloride from Leaves in Carrizo Citrange Seedlings. J. Plant Physiol. 2016, 205, 1–10.
- Bhatta, M.; Morgounov, A.; Belamkar, V.; Wegulo, S.N.; Dababat, A.A.; Erginbas-Orakci, G.; Bouhssini, M.E.; Gautam, P.; Poland, J.; Akci, N.; et al. Genome-Wide Association Study for Multiple Biotic Stress Resistance in Synthetic Hexaploid Wheat. Int. J. Mol. Sci. 2019, 20, 3667.
- Yao, Y.; Carretero-Paulet, L.; Van de Peer, Y. Using Digital Organisms to Study the Evolutionary Consequences of Whole Genome Duplication and Polyploidy. PLoS ONE 2019, 14, e0220257.
- Salman-Minkov, A.; Sabath, N.; Mayrose, I. Whole-Genome Duplication as a Key Factor in Crop Domestication. Nat. Plants 2016, 2, 16115.
- Glombik, M.; Bačovský, V.; Hobza, R.; Kopecký, D. Competition of Parental Genomes in Plant Hybrids. Front. Plant Sci. 2020, 11, 200.
- Guo, H.; Mendrikahy, J.N.; Xie, L.; Deng, J.; Lu, Z.; Wu, J.; Li, X.; Shahid, M.Q.; Liu, X. Transcriptome Analysis of Neo-Tetraploid Rice Reveals Specific Differential Gene Expressions Associated with Fertility and Heterosis. Sci. Rep. 2017, 7, 40139.
- Julião, S.A.; do Ribeiro, C.V.; Lopes, J.M.L.; de Matos, E.M.; Reis, A.C.; Peixoto, P.H.P.; Machado, M.A.; Azevedo, A.L.S.; Grazul, R.M.; de Campos, J.M.S.; et al. Induction of Synthetic Polyploids and Assessment of Genomic Stability in Lippia Alba. Front. Plant Sci. 2020, 11, 292.
- Zhou, L.; Gui, J. Natural and Artificial Polyploids in Aquaculture. Aquac. Fish. 2017, 2, 103–111.
- Vinogradov, A.E.; Borkin, L.J.; Günther, R.; Rosanov, J.M. Two Germ Cell Lineages with Genomes of Different Species in One and the Same Animal. Hereditas 1991, 114, 245–251.
- Vinogradov, A.E.; Borkin, L.J.; Günther, R.; Rosanov, J.M. Genome Elimination in Diploid and Triploid Rana Esculenta Males: Cytological Evidence from DNA Flow Cytometry. Genome 1990, 33, 619–627.
More
