Grain size is a quantitative trait that is controlled by multiple genes. It is not only a yield trait, but also an important appearance quality of rice. In addition, grain size is easy to be selected in evolution, which is also a significant trait for studying rice evolution. In recent years, many quantitative trait loci (QTL)/genes for rice grain size were isolated by map-based cloning or genome-wide association studies, which revealed the genetic and molecular mechanism of grain size regulation in part.
- grain size
- grain width
- grain length
- QTLs
- genes
- rice
1. Introduction
Rice, one of the most important staple foods in the world, is a model plant for knowing plant functional genetics. However, it is still urgent to improve rice grain yield with the continuous increase in the world’s population, the deterioration in the environment, and the decrease in the area of arable land. Rice grain yield is composed of three major factors, including effective panicles per plant, grain number per panicle and 1000-grain weight. The 1000-grain weight is affected by grain shape, grain size and the filling of kernels [1]. Grain size, as specified by its three-dimensional structure of seeds (length, width and thickness), is a key determinant of both yield and appearance quality in rice [2]. In recent years, many quantitative trait loci (QTL)/genes for grain size were isolated in rice by map-based cloning and genome-wide association study (GWAS) and revealed the genetic and molecular mechanisms of grain size regulation in part [3][4][5][6][7][8][9][10][11][12][13][14][15][16][17][18][19][20][21][22][23][24]. These QTL/genes were involved in multiple signaling pathways, including G protein signaling, the mitogen-activated protein kinase (MAPK) signaling pathway, the ubiquitin–proteasome pathway, phytohormone signaling, transcriptional regulatory factors and so on. [25][26]. At present, grain size has become an important target trait for the high-yield and high-quality molecular design breeding of rice. Therefore, summarizing the QTL/genes and analyzing the molecular regulatory pathways of grain size can provide an important theoretical foundation for rice breeding.
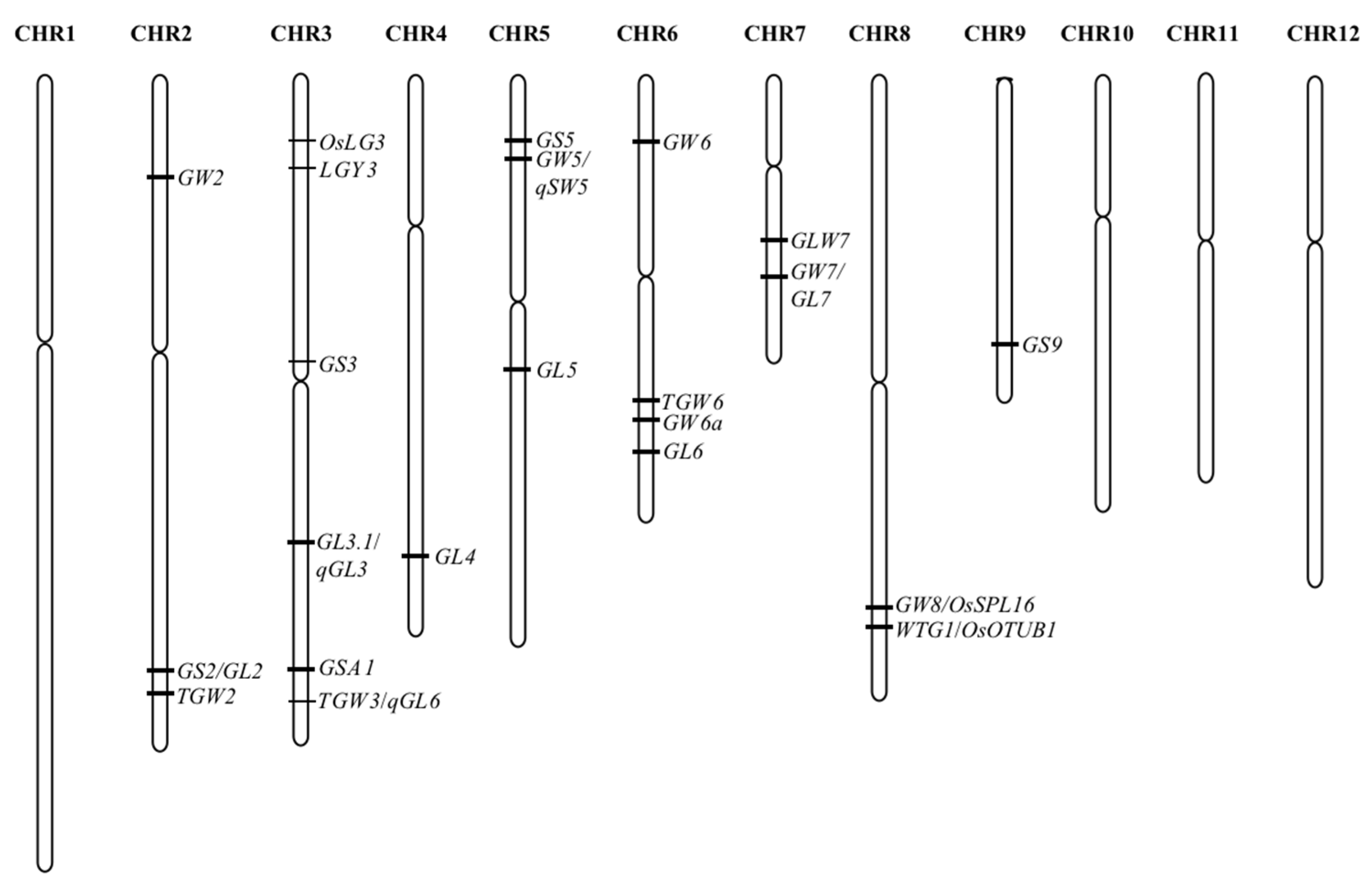
2. Research Progress on the QTL/Genes of Grain Size in Rice
| QTL | Gene ID | Main Regulatory Traits | Signalling Pathways | References |
|---|---|---|---|---|
| GW2 | Os02g0244100 | Width | Ubiquitin–proteasome pathway | [3] |
| GS2 | Os02g0701300 | Size | Phytohormone signaling and transcriptional regulatory factor | [12] |
| TGW2 | Os02g0763000 | Width | Unclear | [23] |
| OsLG3 | Os03g0183000 | Length | Phytohormone signaling and transcriptional regulatory factor | [27] |
| LGY3 | Os03g0215400 | Size | Transcriptional regulatory factor | [28] |
| GS3 | Os03g0407400 | Width | G protein signaling | [4] |
| qGL3 | Os03g0646900 | Size | Phytohormone signaling | [6,8,29][6][8][29] |
| GSA1 | Os03g0757500 | Size | Phytohormone signaling | [30] |
| qTGW3 | Os03g0841800 | Size | Phytohormone signaling | [18] |
| GL4 | ORGLA04G0254300 | Length | Transcriptional regulatory factor | [31] |
| GS5 | Os05g0158500 | Size | Phytohormone signaling | [5] |
| GW5 | Os05g0187500 | Width | Phytohormone signaling and ubiquitin–proteasome pathway | [17,26][17][26] |
| GL5 | Os05g0447200 | Length | Phytohormone signaling | [24] |
| GW6 | Os06g0266800 | Width and length | Phytohormone signaling | [32] |
| TGW6 | Os06g0623700 | Length | Phytohormone signaling | [9] |
| GW6a | Os06g0650300 | Length | transcriptional regulatory factor | [13] |
| GL6 | Os06g0666100 | Length | unclear | [33] |
| GLW7 | Os07g0505200 | Length | transcriptional regulatory factor | [16] |
| GW7/GL7 | Os07g0603300 | Width | transcriptional regulatory factor | [14] |
| GW8 | Os08g0531600 | Width | transcriptional regulatory factor | [14] |
| WTG1 | Os08g0537800 | Width and thickness | Ubiquitin–proteasome pathway | [34,35][34][35] |
| GS9 | Os09g0448500 | Length | transcriptional regulatory factor | [21] |
2.1. QTLs Mainly Associated with Grain Length
2.2. QTLs Mainly Associated with Grain Width
2.3. QTLs Mainly Associated with Thickness
This entry is adapted from 10.3390/ijms23063169
