The estrogen receptor (ER) is the main downstream effector of its ligand estrogen and has functions connected to the menstrual cycle, pregnancy, and lactation in females and in maintaining cardiovascular, nervous, musculoskeletal, and immune system functioning [
The estrogen receptor (ER) is the main downstream effector of its ligand estrogen and has functions connected to the menstrual cycle, pregnancy, and lactation in females and in maintaining cardiovascular, nervous, musculoskeletal, and immune system functioning [
1
]. There are two subtypes of ER, namely ERα and ERβ, encoded by
ESR1
and ESR2, respectively. These genes are located on separate chromosomes,
ESR1
at position 6q24-27 and ESR2 at 14q22-24 [
2
,
3
,
4
]. Whilst highly similar, their ligand binding domains (LBD) differ, enabling specific physiological functions [
1
,
5
,
6].
].
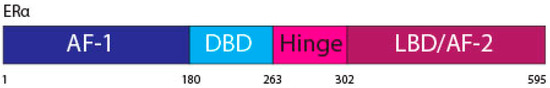
The ERα, a 66 kDa protein, is comprised of four domains, namely activation function 1 (AF-1), DNA binding domain (DBD), the hinge region, and the LBD also known as AF-2 [
The ERα, a 66 kDa protein, is comprised of four domains, namely activation function 1 (AF-1), DNA binding domain (DBD), the hinge region, and the LBD also known as AF-2 [
1
,
7
] (
).
Figure 1.
ERα functional domains. The ERα functional domains include; Activation Function 1 (AF-1) (purple), DNA Binding Domain (DBD) (blue), hinge (pink), Ligand Binding Domain (LBD) and AF-2 (plum).
Upon estrogen binding, ER forms homodimers (ERα/ERα or ERβ/ERβ) and heterodimers (ERα/ERβ) [
Upon estrogen binding, ER forms homodimers (ERα/ERα or ERβ/ERβ) and heterodimers (ERα/ERβ) [
8
,
9
]. This review focuses on
ESR1
encoding ERα, referred to as ER henceforth. Following dimerisation, ER translocates to the nucleus and regulates transcription through estrogen response element (ERE) binding within target gene promoters (
, “genomic function”).
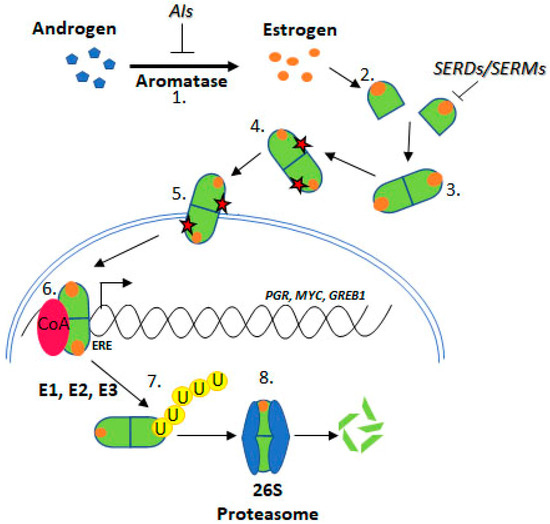
Figure 2. Estrogen Receptor Signalling and ER Targeting Treatment. Aromatase converts androgens to estrogens, Estrogen receptor binds estrogen, Dimerisation, Post translational modification, Nuclear localisation, Coactivator binding and target gene (PGR, c-Myc, GREB1) transcription, Addition of ubiquitin by E1, E2 & E3 ligases, Degradation by the 26S proteasome. Aromatase Inhibitors (AIs) inhibit the enzyme aromatase, Selective Estrogen Receptor Degraders (SERDs) and Selective Estrogen Receptor Modifiers (SERMs) prevent estrogen binding.
Estrogen Receptor Signalling and ER Targeting Treatment. Aromatase converts androgens to estrogens, Estrogen receptor binds estrogen, Dimerisation, Post translational modification, Nuclear localisation, Coactivator binding and target gene (PGR, c-Myc, GREB1) transcription, Addition of ubiquitin by E1, E2 & E3 ligases, Degradation by the 26S proteasome. Aromatase Inhibitors (AIs) inhibit the enzyme aromatase, Selective Estrogen Receptor Degraders (SERDs) and Selective Estrogen Receptor Modifiers (SERMs) prevent estrogen binding.
Alternatively, ER may crosstalk with the PI3K-AKT-mTOR or MAPK pathways, in a “non-genomic” manner (see below), in both scenarios promoting cell proliferation and suppressing apoptosis [
Alternatively, ER may crosstalk with the PI3K-AKT-mTOR or MAPK pathways, in a “non-genomic” manner (see below), in both scenarios promoting cell proliferation and suppressing apoptosis [
10].
].
Approximately 75% of BCa express ER, which promotes oncogenesis. As such, the ER is a common target for BCa treatment using endocrine therapies (ETs) including tamoxifen, fulvestrant, and aromatase inhibitors (AIs) [
Approximately 75% of BCa express ER, which promotes oncogenesis. As such, the ER is a common target for BCa treatment using endocrine therapies (ETs) including tamoxifen, fulvestrant, and aromatase inhibitors (AIs) [
11
] (
). Tamoxifen is known as a selective ER modifier (SERM) and fulvestrant as a selective ER degrader (SERD) [
12
]. SERMs competitively bind ER forming an inactive complex, by preventing coactivator interactions [
12
,
13
,
14
,
15
]. SERDs also competitively bind ER; however, their binding targets the receptor for proteasomal degradation [
16
,
17
,
18
]. AIs are a group of drugs that prevent the synthesis of the estrogen, through inhibition of aromatase [
19
,
20
,
21]. AIs include letrozole, anastrozole, and exemestane. Whilst these treatments are initially effective for many ER positive BCa patients, resistance remains a significant issue.
]. AIs include letrozole, anastrozole, and exemestane. Whilst these treatments are initially effective for many ER positive BCa patients, resistance remains a significant issue.
Resistance to ET is common, approximately 30% of BCa patients acquire resistance [
Resistance to ET is common, approximately 30% of BCa patients acquire resistance [
22
,
23
].
ESR1
mutations have been identified in ET resistant BCa tumours. Several
ESR1
mutations have been functionally characterised and confer key attributes associated with ET resistance, indicating mechanistic roles in resistance such as estrogen independence, increased transcription of ER target genes like PGR (progesterone receptor), GREB1 (growth regulation by estrogen in breast cancer 1), and MYC (c-myc), and increased proliferation and altered ER conformation [
24
,
25
] (
).
Since a major control of ER activity is through regulation of its half-life, changes to ER stability may influence sensitivity to ET. For example, posttranslational modifications (PTMs) and
Since a major control of ER activity is through regulation of its half-life, changes to ER stability may influence sensitivity to ET. For example, posttranslational modifications (PTMs) and
ESR1 mutations causing amino acid substitutions at PTM sites may influence ER stability, and ultimately activity.
mutations causing amino acid substitutions at PTM sites may influence ER stability, and ultimately activity.
2. Estrogen Receptor Signaling
The estrogen/ER complex can result in activation of two distinct types of signaling pathways, the genomic and non-genomic. In the genomic pathway, the ER regulates gene transcription through direct binding of its DBD to promotors containing an ERE, or through interactions with other transcription factors at promoter regions [
The estrogen/ER complex can result in activation of two distinct types of signaling pathways, the genomic and non-genomic. In the genomic pathway, the ER regulates gene transcription through direct binding of its DBD to promotors containing an ERE, or through interactions with other transcription factors at promoter regions [
9
,
26
]. The non-genomic pathway, however, enables rapid signaling through bidirectional crosstalk with PI3K-AKT-mTOR and MAPK pathways. These pathways are frequently upregulated in BCa and enable signaling in an estrogen independent manner [
27
,
28
,
29
]. Several kinases from these pathways phosphorylate the ER at various sites, mediating ER stability, localization, and transactivational capacity, discussed in detail below [
30
,
31
]. Further, these pathways may contribute to ET resistance through phosphorylation at key sites, even in the absence of estrogen [
32
,
33
,
34
,
35
,
36
,
37
]. Additionally, crosstalk between ER and PI3K-AKT-mTOR and MAPK pathways is bidirectional. The ER may activate these pathways through interaction with modulator of non-genomic action of estrogen receptor (MNAR) scaffold protein and subsequent SRC activation [
38
,
39
]. Ultimately, these pathways promote cell cycle progression through increased expression of cyclin D1, and suppress apoptosis [
38
,
40
]. Understanding these interactions is important as the PI3K and MAPK pathways are frequently active in ET resistant BCa and contribute to ET resistance [
27
,
28
,
29
].
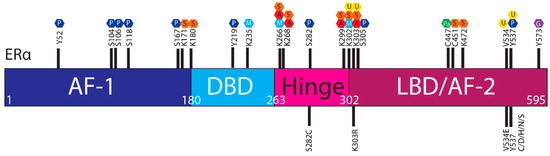
The ER is regulated by a range of PTMs including ubiquitylation, SUMOylation, phosphorylation, palmitoylation, acetylation, methylation and glycosylation [
The ER is regulated by a range of PTMs including ubiquitylation, SUMOylation, phosphorylation, palmitoylation, acetylation, methylation and glycosylation [
41
,
42
,
43
,
44
]. These modifications are proposed to regulate the activity, stability, and interactions of ER with other proteins or DNA, and
ESR1
mutations may influence PTMs and hence ER stability and function (
).
).
Figure 3.
Post translational modifications of ERα. The ERα functional domains include: Activation Function 1 (AF-1) (purple), DNA Binding Domain (DBD) (blue), hinge (pink), Ligand Binding Domain (LBD) and AF-2 (plum). At the top of the schematic are known sites PTMs of ERα; phosphorylation (P, dark blue), SUMOylation (S, orange), methylation (M, light blue), acetylation (A, red), ubiquitylation (U, yellow), pamiltoylation (Pa, green) and gycosylation (G, purple). At the bottom are PTM sites affected by
ESR1
mutations.
2.1. Estrogen Receptor Turnover
Whilst ER is produced through transcription and protein synthesis, changes to ER degradation kinetics is the major factor determining ER levels. In BCa, there is an imbalance between the rate of transcription, synthesis and degradation of the ER leading to increased ER stability and thus activity [
Whilst ER is produced through transcription and protein synthesis, changes to ER degradation kinetics is the major factor determining ER levels. In BCa, there is an imbalance between the rate of transcription, synthesis and degradation of the ER leading to increased ER stability and thus activity [
45
]. The half-life of the ER differs significantly depending on estrogen exposure. Generally, the ER half-life is 3–5 h [
46
] and estrogen presence can reduce the half-life to just 1 h [
47
]. However, persistent estrogen exposure causes relative ER stability. In fact, in MCF-7 cells exposed to estrogen for 72 h, ER half-life was increased to 6 h, due to the decreased rate of proteolysis associated with p-S118 [
48
].
2.2. Degradation by the Ubiquitin Proteasome Pathway
Several studies have shown increased ER stability in the presence of the proteasome inhibitor, MG132, indicating that degradation by the ubiquitin–proteasome system (UPS) regulates ER stability [
Several studies have shown increased ER stability in the presence of the proteasome inhibitor, MG132, indicating that degradation by the ubiquitin–proteasome system (UPS) regulates ER stability [
47
,
49
,
50
,
51]. Degradation of the ER occurs predominantly through the UPS, which relies on ER ubiquitylation by ubiquitin activating enzymes (E1) and ubiquitin conjugating enzymes (E2). Conversely, the ER may be protected from degradation through the activity of ubiquitin ligases (E3), which remove ubiquitin from the ER.
]. Degradation of the ER occurs predominantly through the UPS, which relies on ER ubiquitylation by ubiquitin activating enzymes (E1) and ubiquitin conjugating enzymes (E2). Conversely, the ER may be protected from degradation through the activity of ubiquitin ligases (E3), which remove ubiquitin from the ER.
Ubiquitin conjugation or removal occurs on lysine residues, and mutations resulting in an exchange, either of such lysines or of amino acids affecting the accessibility of important lysines, may therefore alter the protein’s stability. Additionally, some UPS proteins act as coactivators of nuclear receptors and promote downstream gene transcription [
16
,
44
,
52,
,53]. For example, ubiquitin ligase E3A (E6-AP) acts as an ER coactivator and increases gene transcription [16, 53]. For example, ubiquitin ligase E3A (E6-AP) acts as an ER coactivator and increases gene transcription [16][53]
].