Viroids are a group of infectious plant lncRNAs that are composed of RNA genomes and replicate by using the host enzymatic activities.
- Viroids
- circular lncRNAs
- RNA dependent RNA polymerase
- Nuclear encoded polymerase
- Rolling circle replication
- RNA silencing
- Ribozyme
- lncRNAs
- Transposable elements
- Retrotransposons
Definition
Viroids are a group of infectious plant lncRNAs that are composed of RNA genomes and replicate by using the host enzymatic activities.
1. Introduction
Viroids carry the circular, single-stranded, non-coding RNA molecules that represent the smallest known replicons among all living objects, ranging between 246 to 401 nucleotides (nt) in length. Viroid RNAs (vd-RNAs) do not carry any functional open reading frames and can be defined as circular lncRNAs. Their mode of action along with the host processes makes the viroids unusually complex lncRNA molecules.
2. Classification
Viroids can multiply and accumulate in the infected plant tissue by using host RNA polymerases. So far, no animal viroid RNAs have been described. Interestingly, circular single-stranded (ss) viroid RNAs replicate autonomously in plant cellular organelles, either in nuclei or in chloroplasts. No sequence homology with the host plant genome or with a potential helper virus genome has been observed among viroids. Currently, several thousand variants of viroid sequences are available in the National Centre for Biotechnology Information (NCBI) databank. Based on the presence of the central conserved region (CCR) containing the C- domain, viroids are classified into two families, the
Pospiviroidae
and the
Avsunviroidae
, each further divided into genera according to their RNA structures and relationships. A brief overview is given in Table 1.
Table 1.
A brief overview of the various members of viroids belonging to
Avsunviroidae
and
Pospiviroidae
.
|
Viroid Classification |
Type Species |
Genome and Pathogenesis |
|
Avsunviroidae |
||
|
Avsunviroid |
Avocado Sun Blotch Viroid (ASBvd) |
The viroids in this family have a circular genome of 247–399 nucleotides. Viroids are characterized by a specific central conserved region (CCR) in the RNA and have hammerhead ribozymes (HHR) required for symmetric rolling circle replication in the chloroplast. Most viroids are symptomatic to the host whereas some can be asymptomatic. |
|
Pelamoviroid |
Peach latent mosaic viroid (PLMvd) |
|
|
Elaviroid |
Eggplant latent viroid (ELVd) |
|
|
Pospiviroidae |
||
|
Pospiviroid |
Potato spindle tuber viroid (PSTVd) |
The genome size ranges from 246 to 371 nucleotides. The viroids lack a central conserved region (CCR) and ribozyme activity. Replication in the nucleus by asymmetric rolling circle replication is catalyzed completely by host enzymes. It can infect a wide range of hosts including Solanaceae, Asteraceae, Compositae, and others including various economically important fruit crops like apples, citruses, and some ornamental plants. |
|
Hostuviroid |
Hop stunt viroid (HSV) |
|
|
Cocadviroid |
Coconut cadang-cadang viroid (CCCVd) |
|
|
Apscaviroid |
Apple scar skin viroid (ASSVd) |
|
|
Coleviroid |
Coleus blumei viroid (CBVd) |
|
3. Mode of Replication
Both families of viroids adopt the rolling-circle replication (RCR) mechanism, but the
Pospiviridae
replicate by asymmetric and the
Avsunviridae
by symmetric rolling-circle mechanisms (Figure 1).
Pospiviroidae
replicates in the nucleus, where the host DNA dependent RNA polymerase II is involved in the transcription of the viroid RNA. Studies of the PSTVd, the type species of the
Pospiviroidae,
have shown that the host transcription factor TFIIIA is required for the transcription of the viroid RNA. Out of two splicing variants, TFIII-9ZF and TFIII-7ZF, the latter is involved in the transcription of the viroid RNA, which is regulated by the ribosomal protein L5 (RPL5). In
Pospiviroidae
the monomeric circular viroid RNAs replicate by the RCR mechanism to produce an oligomeric longer-than-unit strand, that is cleaved into replicative monomeric RNAs by one or more host ribonucleases (RNases) most likely the members of RNase III family. The recognition of the cleavage site by host RNases is determined by the stem-loop structures in the viroid RNAs around the CCR region. Thus produced monomeric linear (ml) RNA serves as a template for the synthesis of the ml (+) strand RNA. The ml (+) RNA is then ligated into the circular (+) RNA. It has been demonstrated by Nohales et al. (2012) that the viroids force host DNA-ligase, most likely DNA-ligase I, to act as RNA ligase to circularize the ml
(+) strand RNA.
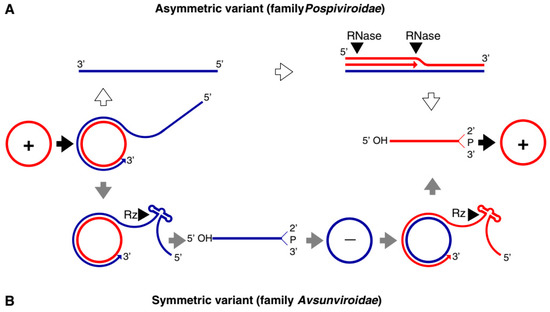
Figure 1.
Rolling circle replication (RCR) in the
Pospiviroidae
and the
Avsunviroidae
. The members of
Pospiviroidae
replicate by the asymmetric RCR mechanism (
A
) inside the host nucleus with the help of the host enzymes. The circular + strand (Red) of viroid RNA is copied into the –strand (Blue) by the RCR, which is linearized by the host RNases, and the linear RNA is then used as the template for the synthesis of the + strand. Finally, the host ligases circularize the viroid RNAs. As shown in panel (
B
), the members of
Avsunviroidae
replicate by the symmetric rolling circle mechanism using the nuclear-encoded polymerase (NEP) in the chloroplast. Replication of RNA strands with both polarities are completed by the RCR mechanism. In this family, the viroid RNAs have autocatalytic ribozymes that catalyze the cleavage of the circular oligomeric form of the viroid RNAs into monomers (Diagram adapted from the Flores et al. FEBS Letters 567, 2004).
The members of
Avsunviroidae
replicate in the chloroplasts by the symmetric RCR mechanism, where (+) stranded circular viroid RNAs are copied into the oligomeric (-) RNAs that are cleaved and ligated to the mc
(-) strands. The mc
(-) strands are used as a template to undergo a second round of the RCR to produce
mc
(+) RNA [17]. The nuclear-encoded polymerase (NEP) are predicted to catalyze the transcription of the viroid RNA at a specific initiation site, which is located at the (A+U)-rich terminal loops in the avocado sun blotch viroid (ASBVd), and a 6–7 bp GUC-rich double-stranded RNA motif in the peach latent mosaic viroid (PLMVd). The cleavage of the replication intermediates of both the polarities required for the replication is known to be facilitated by the autocatalytic hammerhead ribozymes (HHR) in
Avsunviroidae.
Besides, the HHR can also catalyze the ligation reaction in-vitro to circularize the ml
RNAs. However, the efficiency of the HHR and the requirement for a higher concentration of Mg
2+
beyond physiological conditions questioned the feasibility of the reaction in vivo. However, Nohales et al. (2012) have shown that in the replication of eggplant latent viroid (ELVd) the catalytic activity of the chloroplastic isoform of eggplant tRNA ligase mediates the circularization of both ml
(+) and (-) strands. Another work also demonstrated that a recombinant eggplant tRNA ligase can mediate circularization in ASBVd, PLMVd, and Chrysanthemum chlorotic mottle viroid (CChMVd).
The extensive secondary structure plays a critical role in the viroid life cycle, such as in host plant invasion, replication, pathogenesis, and transport. Viroid RNA structures have been studied in great detail by using SHAPE analysis whereas their structural 3D complexity has been confirmed via the direct visualization of single-RNA molecules by atomic force microscopy[1][2]. Structure prediction of the stability of viroid molecules can help explain how viroids escape the RNA silencing pathways[3][4].
The extensive secondary structure plays a critical role in the viroid life cycle, such as in host plant invasion, replication, pathogenesis, and transport. Viroid RNA structures have been studied in great detail by using SHAPE analysis whereas their structural 3D complexity has been confirmed via the direct visualization of single-RNA molecules by atomic force microscopy [31,32]. Structure prediction of the stability of viroid molecules can help explain how viroids escape the RNA silencing pathways [33,34].
4. Infection and Diagnostics
Viroid infections often cause disease symptoms in such important crops as apple, avocado, coconut, grapevine, hop, peach, potato, tomato, and others[5]. Recent findings shed new light on molecular mechanisms of interaction, securing regulation of viroid replication in the plant cell. For example, potato spindle tuber viroid (PSTVd), a model viroid, requires a splicing form of transcription factor IIIA (TFIIIA-7ZF) for its multiplication via direct interaction with a splicing regulator RPL5, which in turn favors the expression of TFIIIA-7ZF[6]. There is some evidence that viroid replication links to RNA silencing, e.g., for PSTVd that can replicate efficiently in the presence of DCL4, but not of DCL2 host genes[7]. Interestingly, small RNAs originated from PLMVd also appeared to participate in the cleavage of chloroplast mRNA for heat shock protein[3], likely inducing the pathogenic symptoms of viroid infection. Similarly, the miRNA-induced cleavage of the virulence region of PSTVd targeted the pyrophosphatase mRNA. PSTVd also targeted a bromo domain-carrying protein VIRP1 in tomato[8]. PSTVd failed to infect VIRP1-suppressed
Viroid infections often cause disease symptoms in such important crops as apple, avocado, coconut, grapevine, hop, peach, potato, tomato, and others [35]. Recent findings shed new light on molecular mechanisms of interaction, securing regulation of viroid replication in the plant cell. For example, potato spindle tuber viroid (PSTVd), a model viroid, requires a splicing form of transcription factor IIIA (TFIIIA-7ZF) for its multiplication via direct interaction with a splicing regulator RPL5, which in turn favors the expression of TFIIIA-7ZF [18]. There is some evidence that viroid replication links to RNA silencing, e.g., for PSTVd that can replicate efficiently in the presence of DCL4, but not of DCL2 host genes [36]. Interestingly, small RNAs originated from PLMVd also appeared to participate in the cleavage of chloroplast mRNA for heat shock protein [33], likely inducing the pathogenic symptoms of viroid infection. Similarly, the miRNA-induced cleavage of the virulence region of PSTVd targeted the pyrophosphatase mRNA. PSTVd also targeted a bromo domain-carrying protein VIRP1 in tomato [37]. PSTVd failed to infect VIRP1-suppressed
N. benthamiana plants, signifying that this viroid should be viewed as a functional lncRNA[5].
plants, signifying that this viroid should be viewed as a functional lncRNA [38].
Viroids have emerged as productive tools with which to study the interactions not only of their replicable lncRNAs, but also the plethora of functions of host lncRNAs in general [9]. Current research focuses on the transcriptomic analysis of viroid-infected plants to identify e.g., the patterns of viroid-siRNA-induced RNA silencing of host mRNAs. This helps to obtain a holistic picture of viroid-induced regulation or the widespread degradation of the host gene expression. Yet translation also appears to be affected/regulated by viroids[10]. It has been shown that viroid molecules were present in the ribosomal fractions[11] In general, the above and similar findings provide new insights to better understand viroid biology and thus the means for viroid control.
Viroids have emerged as productive tools with which to study the interactions not only of their replicable lncRNAs, but also the plethora of functions of host lncRNAs in general [39]. Current research focuses on the transcriptomic analysis of viroid-infected plants to identify e.g., the patterns of viroid-siRNA-induced RNA silencing of host mRNAs. This helps to obtain a holistic picture of viroid-induced regulation or the widespread degradation of the host gene expression. Yet translation also appears to be affected/regulated by viroids [40]. It has been shown that viroid molecules were present in the ribosomal fractions [41]. In general, the above and similar findings provide new insights to better understand viroid biology and thus the means for viroid control.
Among several hypotheses regarding the origination of viroids, Kiefer et al. (1983) suggested that they might come from transposons or retroviruses[12]. However, more currently, because they contain catalytic RNA elements, Flores et al. (2014) considered viroids as remnants of ‘the RNA world’ that arose earlier than DNA and proteins. In some retrotransposons ribozyme activities, were found, further emphasizing a link between TEs and catalytic RNAs[13]. This is supported further by recent findings with ASVd, another model viroid, which has been shown to bear a double hammerhead ribozyme, also found in mobile elements and other viroid-like RNAs [44,45]. Recently, Catalan et al. (2019) have proposed a possibility of the
Among several hypotheses regarding the origination of viroids, Kiefer et al. (1983) suggested that they might come from transposons or retroviruses [42]. However, more currently, because they contain catalytic RNA elements, Flores et al. (2014) considered viroids as remnants of ‘the RNA world’ that arose earlier than DNA and proteins. In some retrotransposons ribozyme activities, were found, further emphasizing a link between TEs and catalytic RNAs [43]. This is supported further by recent findings with ASVd, another model viroid, which has been shown to bear a double hammerhead ribozyme, also found in mobile elements and other viroid-like RNAs [44,45]. Recently, Catalan et al. (2019) have proposed a possibility of the
de novo origin of viroid-like replicons via a parsimonious scenario. From the pool of various RNAs in eukaryotic cells, some can circularize and serve as seeds of the process[14].
origin of viroid-like replicons via a parsimonious scenario. From the pool of various RNAs in eukaryotic cells, some can circularize and serve as seeds of the process [46].
NGS has been widely used in viroid research for the discovery of new viroids and diagnostic purposes. The technology has been employed to analyze viroid sequences and host gene expression (transcriptomic) in response to viroid infection[15]. Progressive filtering of overlapping small RNAs-1/-2 (PROF1/PROF2) uses deeply sequenced small RNAs and assembles them into circular RNAs representing possible viroids and satellite RNAs. This software discovered the apple hammerhead viroid (AHVd) from apple plants and a novel grapevine latent viroid (GLVd) from an old grapevine plant[16][17]. Recently, the PROF2 in combination with the assembly software Velvet allowed the discovery of viroid-derived small RNAs responsible for host RNA silencing in
NGS has been widely used in viroid research for the discovery of new viroids and diagnostic purposes. The technology has been employed to analyze viroid sequences and host gene expression (transcriptomic) in response to viroid infection [47]. Progressive filtering of overlapping small RNAs-1/-2 (PROF1/PROF2) uses deeply sequenced small RNAs and assembles them into circular RNAs representing possible viroids and satellite RNAs. This software discovered the apple hammerhead viroid (AHVd) from apple plants and a novel grapevine latent viroid (GLVd) from an old grapevine plant [48]. Recently, the PROF2 in combination with the assembly software Velvet allowed the discovery of viroid-derived small RNAs responsible for host RNA silencing in
Coleus blumei infected with coleus blumei viroids (CbVds). Based on the NGS data it has been revealed that the central conserved region (CCR) of the viroid is pivotal in the biogenesis of sRNAs for both host RNA silencing and the genome replication of the viroids [18] Deep sequencing revealed the viroid heterogeneity, with 3939 variants of inoculated parent PLMVd detected after six months of infection in the natural host[19]. Similarly, transcriptome analysis of the citrus bark cracking viroid (CBCVd) revealed a new variant of the viroid from citrus plants in Pakistan. Interestingly, the viroid was highly diverse phylogenetically from CBCVd found in other Asian countries[17]. Therefore, NGS has proved to be an important asset in the discovery of novel viroids and can be used for viroid screening and quarantine.
infected with coleus blumei viroids (CbVds). Based on the NGS data it has been revealed that the central conserved region (CCR) of the viroid is pivotal in the biogenesis of sRNAs for both host RNA silencing and the genome replication of the viroids [49]. Deep sequencing revealed the viroid heterogeneity, with 3939 variants of inoculated parent PLMVd detected after six months of infection in the natural host [50]. Similarly, transcriptome analysis of the citrus bark cracking viroid (CBCVd) revealed a new variant of the viroid from citrus plants in Pakistan. Interestingly, the viroid was highly diverse phylogenetically from CBCVd found in other Asian countries [51]. Therefore, NGS has proved to be an important asset in the discovery of novel viroids and can be used for viroid screening and quarantine.
