Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Bruce Ren and Version 1 by Mabel Buelna-Chontal.
The transcription factor Nrf2 is a master regulator of multiple cytoprotective genes that maintain redox homeostasis and exert anti-inflammatory functions. The Nrf2-Keap1 signaling pathway is a paramount target of many cardioprotective strategies, because redox homeostasis is essential in cardiovascular health. Nrf2 gene variations, including single nucleotide polymorphisms (SNPs), are correlated with cardiometabolic diseases and drug responses. SNPs of Nrf2, KEAP1, and other related genes can impair the transcriptional activation or the activity of the resulting protein, exerting differential susceptibility to cardiometabolic disease progression and prevalence.
- redox control
- Nrf2
- single nucleotide polymorphisms
- cardiometabolic diseases
1. Introduction
Worldwide cardiometabolic diseases-related deaths have been increasing over the last decades, representing a big concern [1]. As cardiometabolic diseases lead to unbalance of redox homeostasis and enhanced oxidative stress (for a recent comprehensive review, see [2]), different antioxidant therapies have been evaluated to prevent and treat such diseases. Although some of these have been designed to induce the nuclear factor erythroid 2-related factor 2 (Nrf2)-driven antioxidant response [3], their efficacy in the clinic has not been clearly established, giving rise to the possibility of the participation of other regulatory elements in the Nrf2-signaling cascade. In this regard, multiple single nucleotide polymorphisms (SNPs) have been found in the Nrf2 gene [4]. Overall, SNPs of the Nrf2 gene induce changes in Nrf2 activation, which can lead to the impairment of the endogenous antioxidant system, contributing to chronic inflammation and other detrimental effects [5]. Indeed, several polymorphisms of Nrf2 have been associated with cardiometabolic diseases progression [6].
2. The Role of Nrf2 to Maintain Redox Homeostasis in Cardiometabolic Diseases
A common feature of cardiometabolic diseases is the imbalance between pro- and anti-oxidative factors in the cell. Such condition is associated with high levels of reactive oxygen species (ROS) and reactive nitrogen species (RNS) that react with lipids, proteins, DNA, or activate redox signaling pathways, leading to cellular injury and death. Disruption of the redox homeostasis is associated with poor cardiac contractility related to reduced sarco-endoplasmic reticulum Ca2+-ATPase (SERCA2A) activity [7], but also with mitochondrial damage. Mitochondria are the primary ROS producers and, at the same time, are the first exposed in front line due to their deleterious action, resulting in further mitochondrial dysfunction and a vicious cycle of ROS generation. Impairment of the mitochondrial function and increased rates of fatty acid β-oxidation promote the accumulation of incomplete β-oxidation products, which, in combination with oxidative stress, contribute to insulin resistance in cardiometabolic diseases [8].
In cardiometabolic diseases, among the antioxidant molecules regulated by Nrf2 are catalase (CAT), superoxide dismutase (SOD), glutathione S-transferases (GST), glutathione peroxidases (GPx), heme-oxygenase 1 (HMOX-1), thioredoxin (TXN), thioredoxin reductases, peroxiredoxins (Prdx), and NAD(P)H quinone oxidoreductase-1 (NQO1) [9][10][11][12] (Figure 1A). Besides the antioxidant response regulation, Nrf2 also modulates a myriad of genes related to other cellular processes. For instance, it is known that Nrf2 regulates the expression of Notch1 and downstream genes, which collectively coordinate signals that affect differentiation, proliferation, and apoptotic events [13]. Moreover, it was demonstrated by chromatin immunoprecipitation that Nrf2 binds directly to an antioxidant response elements (ARE) region in the aryl hydrocarbon receptor (AHR) promoter [14]. AHR increased in pig myocardium after ischemia-reperfusion damage and diminished upon administration of natural flavone baicalin [15] and sulforaphane [16] in association with lesser cardiac injury and inflammation.
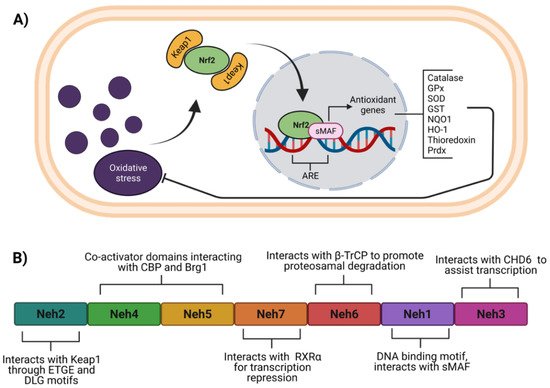

Figure 1. Function and structure of Nrf2. (A) Under oxidative stress, Nrf2 dissociates from Keap1, to be translocated to the nucleus where heterodimerizes with the sMAF proteins for recognition of ARE within several genes to induce their expression. (B) Nrf2 consists of seven domains named Neh1 to 7, which allow specific interaction for its regulation. ARE, antioxidant response elements; β-TrCP, beta-transducin repeat-containing protein; Brg1, Brahma-related gene 1; CBP, CREB binding protein; CDH6, chromo-ATPase/helicase DNA binding protein 6; GPx, glutathione peroxidase; GST, glutathione-S-transferase (GST); HO-1, heme-oxygenase 1; Keap1, Kelch-like ECH-associated protein 1; NQO1, NAD(P)H quinone oxidoreductase 1; Neh, Nrf2-ECH homology; Nrf2, nuclear factor erythroid 2-related factor 2; Prdx, peroxiredoxins; RXRα, retinoid X receptor alpha; SOD, superoxide dismutase; sMAF, small musculoaponeurotic fibrosarcoma. Figure created with BioRender at biorender.com (accessed on 10 February 2022).
Nrf2 Structure and Regulatory Mechanisms
Nrf2 belongs to the cap‘n’ collar (CNC) subfamily of the basic leucine zipper (bZip) transcription factors [17]. Nrf2 contains seven domains (Figure 1B) named Nrf2-ECH homology (Neh) 1 to 7 [18][19]. Among Neh domains, Neh1 and Neh2 have the most recognizable roles in Nrf2 function and regulation. Neh2 domain is located at the amino-terminal and has ETGE and DLG motifs that interact with Kelch-like ECH-associated protein 1 (Keap1), the well-known negative regulator of Nrf2 [20]. Neh4 and Neh5 are two domains that allow Nrf2 interaction with other proteins such as co-activators [21][22][23][24], whereas Neh5 possesses a nuclear export signal (NES) sensitive to oxidation [25]. Neh7 and Neh6 domains act as negative regulators, and Neh7 interacts with the retinoic X receptor alpha (RXRα) to repress Nrf2 transcriptional activity. In fact, RXRα competes with Nrf2 for binding to the ARE sequence [19]. On the other hand, Neh6 interacts with beta-transducin repeat-containing protein (β-TrCP) to promote Nrf2 ubiquitination and consequent degradation [26][27]. Neh1 domain located near the C-terminal region harbors a basic leucine zipper motif, which binds to ARE sequences within target genes. Neh1 also interacts with small musculoaponeurotic fibrosarcoma (sMAF) proteins favoring the transcription of antioxidant genes. In addition, Neh1 acetylation enhances the transcriptional activity of Nrf2 [28][29]. The Neh3 domain in the C-terminal region interacts with chromo-ATPase/helicase DNA binding protein 6 (CDH6), essential for Nrf2 transcriptional activity [30].
Under homeostatic conditions, Nrf2 is maintained at low levels due to constant proteasomal degradation. The degradation process occurs through the interaction of the ETGE and DLG motifs within the Nrf2 Neh2 domain with a Keap1 homodimer [20]. Keap1 acts as an adaptor for an E3 ubiquitin ligase complex that contains cullin 3 (CUL3) and ring box-1 (Rbx) [31], which catalyze the ubiquitination of lysine residues located between ETGE and DLG motifs [32][33] to induce Nrf2 degradation through the ubiquitin proteasomal system (UPS). Interestingly, Nrf2 also promotes CUL3 and Rbx expression as negative feedback [34]. Another mechanism that induces Nrf2 degradation via UPS is Neh6 phosphorylation by glycogen synthase kinase 3 (GSK-3); this promotes the binding of β-TrCP, which acts as a substrate for the Skp1–Cullin–F-box protein complex (SFC), another E3 ubiquitin ligase complex (Figure 2A) [35]. Of note, the levels of GSK-3 increase in experimental models of obesity, diabetes, and associated cardiovascular diseases [36][37][38].
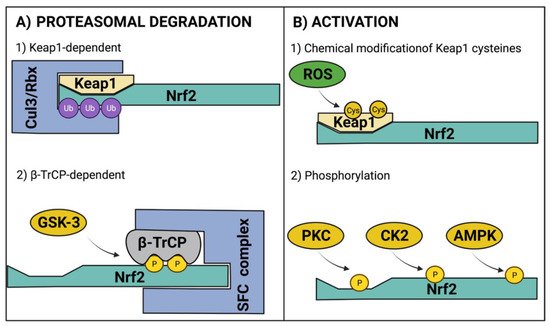

Figure 2. Regulatory mechanisms of Nrf2. (A) For Nrf2 regulation at protein level, Nrf2 is ubiquitinated (Ub) for its proteasomal degradation through Keap1-dependent and β-TrCP-dependent mechanisms. (B) For Nrf2 activation, the canonical pathway involves the chemical modification of Keap1 cysteines (Cys) that elicits the release of Nrf2 for its nuclear translocation. Additionally, phosphorylation (P) by cytosolic kinases promotes Nrf2 activation. AMPK, AMP-activated protein kinase; β-TrCP, beta-transducin repeat-containing protein; CK2, casein kinase 2; Cul3, cullin 3; GSK-3, glycogen synthase kinase 3; Keap1, kelch-like ECH-associated protein 1; Nrf2, nuclear factor erythroid 2-related factor 2; PKC, protein kinase C; Rbx, ring box-1; ROS, reactive oxygen species; SFC, Skp1–Cullin–F-box protein complex. Figure created with BioRender at biorender.com (accessed on 10 February 2022).
During oxidative stress, ROS can react with Keap1 cysteine residues modifying the Keap1 homodimer structure, allowing Nrf2 release. Hence, Keap1 also acts as a sensor of redox homeostasis [39][40][41][42][43]. Once Nrf2 is released from the Keap1/Cul3/Rbx complex, nuclear localization signals (NLS) are exposed for importins recognition to elicit Nrf2 nuclear translocation [44]. After heterodimerization with sMAF, Nrf2 induces the expression of antioxidant response-related genes [45]. Another potential mechanism for Nrf2 activation involves its phosphorylation by several kinases. Protein kinase C (PKC) phosphorylates Nrf2 at serine 40 of the Neh2 domain, inducing its nuclear translocation [46]. Casein kinase 2 (CK2) can phosphorylate Neh4 and Neh5 transactivation domains, enhancing Nrf2 transcriptional activity [47]; in addition, CK2 phosphorylates AMP-activated protein kinase (AMPK) [48], which, in turn, phosphorylate serine residue 558 to increase Nrf2 nuclear accumulation [49], and serine residues 374, 408, 433 to increase its transcriptional activity [50]. The regulatory mechanisms of Nrf2 are depicted in Figure 2.
References
- Cardiometabolic Diseases. 2022. Available online: https://www.who.int/news-room/fact-sheets/detail/cardiovascular-diseases-(cvds) (accessed on 28 January 2022).
- Barteková, M.; Adameová, A.; Görbe, A.; Ferenczyová, K.; Pecháňová, O.; Lazou, A.; Dhalla, N.S.; Ferdinandy, P.; Giricz, Z. Natural and synthetic antioxidants targeting cardiac oxidative stress and redox signaling in cardiometabolic diseases. Free Radic. Biol. Med. 2021, 169, 446–477.
- Patel, B.; Mann, G.E.; Chapple, S.J. Concerted redox modulation by sulforaphane alleviates diabetes and cardiometabolic syndrome. Free Radic. Biol. Med. 2018, 122, 150–160.
- Yamamoto, T.; Yoh, K.; Kobayashi, A.; Ishii, Y.; Kure, S.; Koyama, A.; Sakamoto, T.; Sekizawa, K.; Motohashi, H.; Yamamoto, M. Identification of polymorphisms in the promoter region of the human NRF2 gene. Biochem. Biophys. Res. Commun. 2004, 321, 72–79.
- Hua, C.-C.; Chang, L.-C.; Tseng, J.-C.; Chu, C.-M.; Liu, Y.-C.; Shieh, W.-B. Functional haplotypes in the promoter region of transcription factor Nrf2 in chronic obstructive pulmonary disease. Dis. Markers 2010, 28, 185–193.
- Cho, H.-Y.; Marzec, J.; Kleeberger, S.R. Functional polymorphisms in Nrf2: Implications for human disease. Free Radic. Biol. Med. 2015, 88, 362–372.
- Villalobos, J.B.; Molina-Muñoz, T.; Mailloux-Salinas, P.; Bravo, G.; Carvajal, K.; Gómez-Víquez, N.L. Oxidative stress in cardiomyocytes contributes to decreased SERCA2a activity in rats with metabolic syndrome. Am. J. Physiol. Heart Circ. Physiol. 2013, 305, H1344–H1353.
- Ramírez-Camacho, I.; García-Niño, W.; Flores-García, M.; Pedraza-Chaverri, J.; Zazueta, C. Alteration of mitochondrial supercomplexes assembly in metabolic diseases. Biochim. Biophys. Acta (BBA)-Mol. Basis Dis. 2020, 1866, 165935.
- Pisoschi, A.M.; Pop, A. The role of antioxidants in the chemistry of oxidative stress: A review. Eur. J. Med. Chem. 2015, 97, 55–74.
- Lee, Y.; Im, E. Regulation of miRNAs by Natural Antioxidants in Cardiovascular Diseases: Focus on SIRT1 and eNOS. Antioxidants 2021, 10, 377.
- Hayes, J.D.; Dinkova-Kostova, A. The Nrf2 regulatory network provides an interface between redox and intermediary metabolism. Trends Biochem. Sci. 2014, 39, 199–218.
- Ibrahim, L.; Mesgarzadeh, J.; Xu, I.; Powers, E.T.; Wiseman, R.L.; Bollong, M.J. Defining the Functional Targets of Cap‘n’collar Transcription Factors NRF1, NRF2, and NRF3. Antioxidants 2020, 9, 1025.
- Wakabayashi, N.; Slocum, S.L.; Skoko, J.J.; Shin, S.; Kensler, T.W. When NRF2 Talks, Who’s Listening? Antioxid. Redox Signal. 2010, 13, 1649–1663.
- Shin, S.; Wakabayashi, N.; Misra, V.; Biswal, S.; Lee, G.H.; Agoston, E.S.; Yamamoto, M.; Kensler, T.W. NRF2 Modulates Aryl Hydrocarbon Receptor Signaling: Influence on Adipogenesis. Mol. Cell. Biol. 2007, 27, 7188–7197.
- Xue, Y.; Shui, X.; Su, W.; He, Y.; Lu, X.; Zhang, Y.; Yan, G.; Huang, S.; Lei, W.; Chen, C. Baicalin inhibits inflammation and attenuates myocardial ischaemic injury by aryl hydrocarbon receptor. J. Pharm. Pharmacol. 2015, 67, 1756–1764.
- Silva-Palacios, A.; Ostolga-Chavarría, M.; Sánchez-Garibay, C.; Rojas-Morales, P.; Galván-Arzate, S.; Buelna-Chontal, M.; Pavón, N.; Pedraza-Chaverrí, J.; Königsberg, M.; Zazueta, C. Sulforaphane protects from myocardial ischemia-reperfusion damage through the balanced activation of Nrf2/AhR. Free Radic. Biol. Med. 2019, 143, 331–340.
- Moi, P.; Chan, K.; Asunis, I.; Cao, A.; Kan, Y.W. Isolation of NF-E2-related factor 2 (Nrf2), a NF-E2-like basic leucine zipper transcriptional activator that binds to the tandem NF-E2/AP1 repeat of the beta-globin locus control region. Proc. Natl. Acad. Sci. USA 1994, 91, 9926–9930.
- Itoh, K.; Wakabayashi, N.; Katoh, Y.; Ishii, T.; Igarashi, K.; Engel, J.D.; Yamamoto, M. Keap1 represses nuclear activation of antioxidant responsive elements by Nrf2 through binding to the amino-terminal Neh2 domain. Genes Dev. 1999, 13, 76–86.
- Wang, H.; Liu, K.; Geng, M.; Gao, P.; Wu, X.; Hai, Y.; Li, Y.; Li, Y.; Luo, L.; Hayes, J.; et al. RXRα Inhibits the NRF2-ARE Signaling Pathway through a Direct Interaction with the Neh7 Domain of NRF2. Cancer Res. 2013, 73, 3097–3108.
- Tong, K.I.; Katoh, Y.; Kusunoki, H.; Itoh, K.; Tanaka, T.; Yamamoto, M. Keap1 Recruits Neh2 through Binding to ETGE and DLG Motifs: Characterization of the Two-Site Molecular Recognition Model. Mol. Cell. Biol. 2006, 26, 2887–2900.
- Katoh, Y.; Itoh, K.; Yoshida, E.; Miyagishi, M.; Fukamizu, A.; Yamamoto, M. Two domains of Nrf2 cooperatively bind CBP, a CREB binding protein, and synergistically activate transcription. Genes Cells 2001, 6, 857–868.
- Zhang, J.; Ohta, T.; Maruyama, A.; Hosoya, T.; Nishikawa, K.; Maher, J.M.; Shibahara, S.; Itoh, K.; Yamamoto, M. BRG1 Interacts with Nrf2 to Selectively Mediate HO-1 Induction in Response to Oxidative Stress. Mol. Cell. Biol. 2006, 26, 7942–7952.
- Shen, G.; Hebbar, V.; Nair, S.; Xu, C.; Li, W.; Lin, W.; Keum, Y.-S.; Han, J.; Gallo, M.A.; Kong, A.-N.T. Regulation of Nrf2 Transactivation Domain Activity. The differential effects of mitogen-activated protein kinase cascades and synergistic stimulatory effect of Raf and CREB-binding protein. J. Biol. Chem. 2004, 279, 23052–23060.
- Zhang, J.; Hosoya, T.; Maruyama, A.; Nishikawa, K.; Maher, J.M.; Ohta, T.; Motohashi, H.; Fukamizu, A.; Shibahara, S.; Itoh, K.; et al. Nrf2 Neh5 domain is differentially utilized in the transactivation of cytoprotective genes. Biochem. J. 2007, 404, 459–466.
- Li, W.; Yu, S.; Kong, A.-N.T. Nrf2 Possesses a Redox-sensitive Nuclear Exporting Signal in the Neh5 Transactivation Domain. J. Biol. Chem. 2006, 281, 27251–27263.
- Chowdhry, S.; Zhang, Y.; McMahon, M.; Sutherland, C.; Cuadrado, A.; Hayes, J.D. Nrf2 is controlled by two distinct β-TrCP recognition motifs in its Neh6 domain, one of which can be modulated by GSK-3 activity. Oncogene 2013, 32, 3765–3781.
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J.D. Redox-regulated Turnover of Nrf2 Is Determined by at Least Two Separate Protein Domains, the Redox-sensitive Neh2 Degron and the Redox-insensitive Neh6 Degron. J. Biol. Chem. 2004, 279, 31556–31567.
- Sun, Z.; Chin, Y.E.; Zhang, D.D. Acetylation of Nrf2 by p300/CBP Augments Promoter-Specific DNA Binding of Nrf2 during the Antioxidant Response. Mol. Cell. Biol. 2009, 29, 2658–2672.
- Li, W.; Yu, S.; Liu, T.; Kim, J.-H.; Blank, V.; Li, H.; Kong, A.-N.T. Heterodimerization with small Maf proteins enhances nuclear retention of Nrf2 via masking the NESzip motif. Biochim. Biophys. Acta 2008, 1783, 1847–1856.
- Nioi, P.; Nguyen, T.; Sherratt, P.J.; Pickett, C.B. The Carboxy-Terminal Neh3 Domain of Nrf2 Is Required for Transcriptional Activation. Mol. Cell. Biol. 2005, 25, 10895–10906.
- Kobayashi, A.; Kang, M.-I.; Okawa, H.; Ohtsuji, M.; Zenke, Y.; Chiba, T.; Igarashi, K.; Yamamoto, M. Oxidative Stress Sensor Keap1 Functions as an Adaptor for Cul3-Based E3 Ligase To Regulate Proteasomal Degradation of Nrf2. Mol. Cell. Biol. 2004, 24, 7130–7139.
- McMahon, M.; Thomas, N.; Itoh, K.; Yamamoto, M.; Hayes, J. Dimerization of Substrate Adaptors Can Facilitate Cullin-mediated Ubiquitylation of Proteins by a “Tethering” Mechanism: A two-site interaction model for the Nrf2-Keap1 complex. J. Biol. Chem. 2006, 281, 24756–24768.
- Zhang, D.D.; Lo, S.-C.; Cross, J.V.; Templeton, D.J.; Hannink, M. Keap1 Is a Redox-Regulated Substrate Adaptor Protein for a Cul3-Dependent Ubiquitin Ligase Complex. Mol. Cell. Biol. 2004, 24, 10941–10953.
- Kaspar, J.W.; Jaiswal, A.K. An Autoregulatory Loop between Nrf2 and Cul3-Rbx1 Controls Their Cellular Abundance. J. Biol. Chem. 2010, 285, 21349–21358.
- Rada, P.; Rojo, A.I.; Chowdhry, S.; McMahon, M.; Hayes, J.D.; Cuadrado, A. SCF/-TrCP Promotes Glycogen Synthase Kinase 3-Dependent Degradation of the Nrf2 Transcription Factor in a Keap1-Independent Manner. Mol. Cell. Biol. 2011, 31, 1121–1133.
- Wang, L.; Wang, Y.; Zhang, C.; Li, J.; Meng, Y.; Dou, M.; Noguchi, C.T.; Di, L. Inhibiting Glycogen Synthase Kinase 3 Reverses Obesity-Induced White Adipose Tissue Inflammation by Regulating Apoptosis Inhibitor of Macrophage/CD5L-Mediated Macrophage Migration. Arter. Thromb. Vasc. Biol. 2018, 38, 2103–2116.
- Eldar-Finkelman, H.; Schreyer, S.A.; Shinohara, M.M.; LeBoeuf, R.C.; Krebs, E.G. Increased glycogen synthase kinase-3 activity in diabetes- and obesity-prone C57BL/6J mice. Diabetes 1999, 48, 1662–1666.
- Wu, W.; Liu, X.; Han, L. Apoptosis of cardiomyocytes in diabetic cardiomyopathy involves overexpression of glycogen synthase kinase-3β. Biosci. Rep. 2019, 39, BSR20171307.
- Yamamoto, T.; Suzuki, T.; Kobayashi, A.; Wakabayashi, J.; Maher, J.; Motohashi, H.; Yamamoto, M. Physiological Significance of Reactive Cysteine Residues of Keap1 in Determining Nrf2 Activity. Mol. Cell. Biol. 2008, 28, 2758–2770.
- Takaya, K.; Suzuki, T.; Motohashi, H.; Onodera, K.; Satomi, S.; Kensler, T.W.; Yamamoto, M. Validation of the multiple sensor mechanism of the Keap1-Nrf2 system. Free Radic. Biol. Med. 2012, 53, 817–827.
- McMahon, M.; Lamont, D.J.; Beattie, K.A.; Hayes, J.D. Keap1 perceives stress via three sensors for the endogenous signaling molecules nitric oxide, zinc, and alkenals. Proc. Natl. Acad. Sci. USA 2010, 107, 18838–18843.
- Luo, Y.; Eggler, A.L.; Liu, D.; Liu, G.; Mesecar, A.D.; van Breemen, R.B. Sites of alkylation of human Keap1 by natural chemoprevention agents. J. Am. Soc. Mass Spectrom. 2007, 18, 2226–2232.
- Zhang, D.D.; Hannink, M. Distinct Cysteine Residues in Keap1 Are Required for Keap1-Dependent Ubiquitination of Nrf2 and for Stabilization of Nrf2 by Chemopreventive Agents and Oxidative Stress. Mol. Cell. Biol. 2003, 23, 8137–8151.
- Theodore, M.; Kawai, Y.; Yang, J.; Kleshchenko, Y.; Reddy, S.P.; Villalta, F.; Arinze, I.J. Multiple Nuclear Localization Signals Function in the Nuclear Import of the Transcription Factor Nrf2. J. Biol. Chem. 2008, 283, 8984–8994, Erratum in J. Biol. Chem. 2008, 283, 14176.
- Katsuoka, F.; Motohashi, H.; Ishii, T.; Aburatani, H.; Engel, J.D.; Yamamoto, M. Genetic Evidence that Small Maf Proteins Are Essential for the Activation of Antioxidant Response Element-Dependent Genes. Mol. Cell. Biol. 2005, 25, 8044–8051.
- Bloom, D.A.; Jaiswal, A.K. Phosphorylation of Nrf2 at Ser40 by Protein Kinase C in Response to Antioxidants Leads to the Release of Nrf2 from INrf2, but Is Not Required for Nrf2 Stabilization/Accumulation in the Nucleus and Transcriptional Activation of Antioxidant Response Element-mediated NAD(P)H:Quinone Oxidoreductase-1 Gene Expression. J. Biol. Chem. 2003, 278, 44675–44682.
- Apopa, P.L.; He, X.; Ma, Q. Phosphorylation of Nrf2 in the transcription activation domain by casein kinase 2 (CK2) is critical for the nuclear translocation and transcription activation function of Nrf2 in IMR-32 neuroblastoma cells. J. Biochem. Mol. Toxicol. 2008, 22, 63–76.
- Jang, D.E.; Song, J.; Park, J.-W.; Yoon, S.-H.; Bae, Y.-S. Protein kinase CK2 activates Nrf2 via autophagic degradation of Keap1 and activation of AMPK in human cancer cells. BMB Rep. 2020, 53, 272–277.
- Joo, M.S.; Kim, W.D.; Lee, K.Y.; Kim, J.H.; Koo, J.H.; Kim, S.G. AMPK Facilitates Nuclear Accumulation of Nrf2 by Phosphorylating at Serine 550. Mol. Cell. Biol. 2016, 36, 1931–1942.
- Matzinger, M.; Fischhuber, K.; Pölöske, D.; Mechtler, K.; Heiss, E.H. AMPK leads to phosphorylation of the transcription factor Nrf2, tuning transactivation of selected target genes. Redox Biol. 2020, 29, 101393.
More
