Cells use an impressive array of components to enable the safe transport of protein cargo from the cytosolic ribosomes to the endoplasmic reticulum. Safety during the transit is warranted by the interplay of cytosolic chaperones, membrane receptors, and protein translocases that together form functional networks and serve as protein targeting and translocation routes. While two targeting routes to the endoplasmic reticulum, SRP (signal recognition particle) and GET (guided entry of tail-anchored proteins), prefer targeting determinants at the N- and C-terminus of the cargo polypeptide, respectively, the discovered SND (SRP-independent) route seems to preferentially cater for cargos with non-generic targeting signals that are less hydrophobic or more distant from the termini.
- endoplasmic reticulum
- protein targeting
- protein transport
- Sec61 complex
- EMC
- GET
- SND
- SRP
1. Introduction
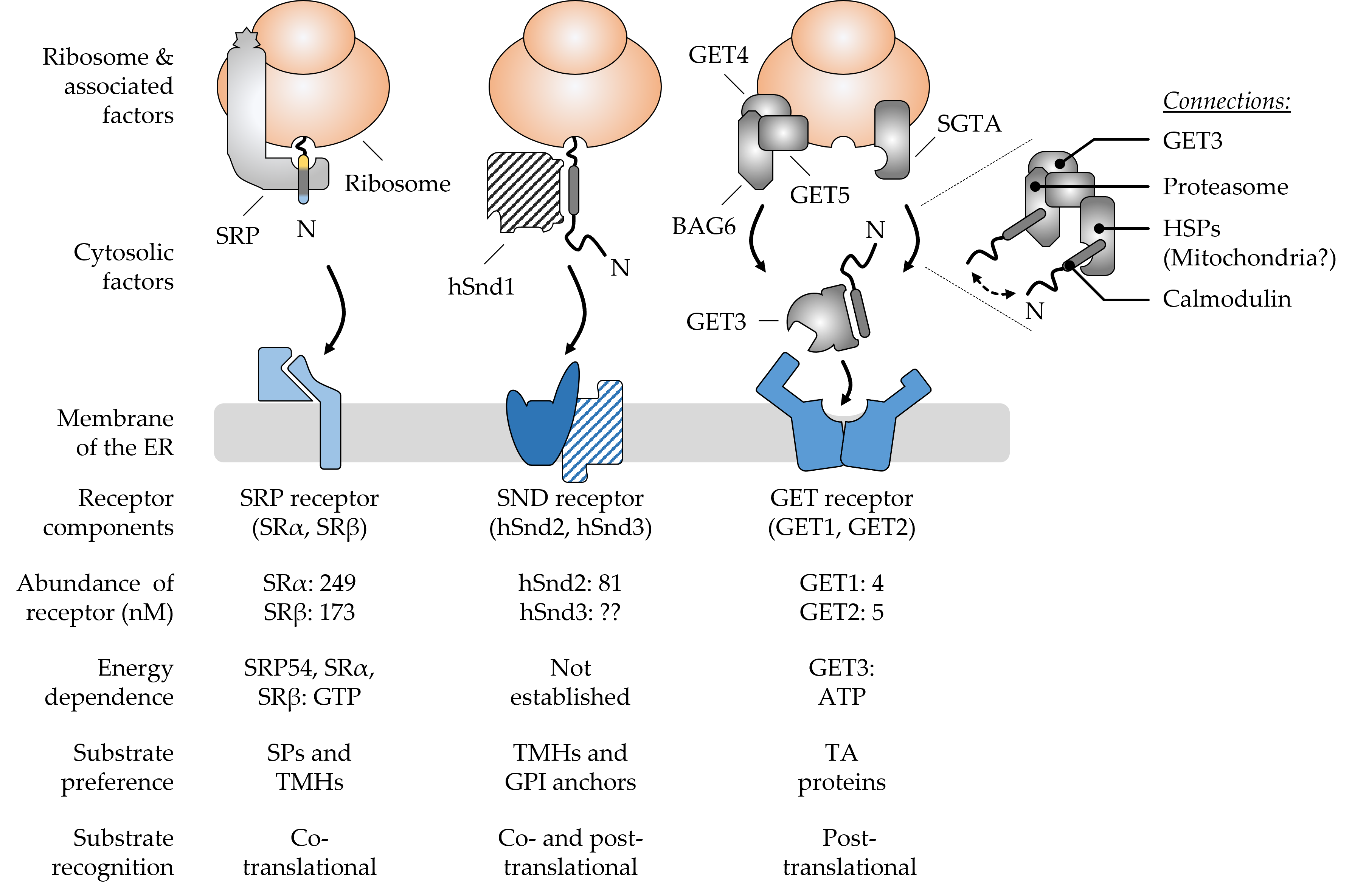
Figure 1. Major components and hallmarks of the mammalian SRP, SND, and GET targeting pathways. The top half shows a graphical output of the major components that shape the three targeting pathways SRP, SND, and GET. The dotted lines indicate a zoomed-in view of the BAG6 pre-targeting complex cooperating with SGTA and other cellular components. The double-headed arrow suggests the cycling of TA proteins between the two chaperones SGTA and BAG6. The bottom half summarizes some of the key features that differentiate the pathways from each other. Ribosome-associated and cytosolic targeting components are shown in grey colors and the cognate membrane receptors in shades of blue. Components (hSnd1, hSnd3) shown with a hatched color fill have not yet been identified in higher eucaryotes. Their existence is based on findings from yeast and the conserved nature of targeting machineries [1]. Abundance values for the receptor components are based on the quantitative mass spectrometry of mammalian cells [2]. Please note, there is considerable controversy about the abundance of GET1 and GET2 with some sources finding GET2 in four- to sevenfold molar excess over GET1 [3][4]. The N-terminus (N) of newly synthesized polypeptides is shown to accentuate the positioning of signal peptides (SPs) and transmembrane helices (TMHs) of different types of cargos, including tail-anchored (TA) proteins and glycosylphosphatidylinositol (GPI)-anchored proteins. BAG6, BCL2-associated athanogene 6; GET, guided entry of TA proteins; HSPs, heat shock proteins; SGTA, small glutamine rich tetratricopeptide repeat co-chaperone alpha; SND, SRP-independent; SR, SRP receptor; SRP, signal recognition particle.
After targeting to the ER membrane, co- and post-translationally arriving polypeptides rely on one of the protein translocation machineries residing in this membrane. Similar to the diversity of precursor polypeptides that are handled by multiple targeting pathways, multiple protein translocases have also evolved to support the insertion or translocation of proteins into or across the ER membrane [26][27]. As the ER membrane originated from the procaryotic plasma membrane, some of the translocation machines of the ER resemble their bacterial ancestors [28][29]. The first ER protein translocase that was described and studied in much detail was the Sec61 complex [30]. This translocase can open up a “fenestrated” conduit through the ER membrane for the translocation of SP-carrying proteins as well as the lateral release of TMHs [30][31]. Other precursor polypeptides, such as TA proteins, can make use of alternative translocation machines such as the GET1/2 complex or the ER membrane protein complex (EMC) [32][33]. Interestingly, precursor proteins relying on the GET1/2 complex or EMC require a differentially shaped opening compared to the membrane-spanning pore that is provided by the Sec61 complex. This shows that nature has apparently found more than one solution for polypeptides to traverse a membrane.2. Different ER Protein Translocases Act as Membrane-Integrated Chaperones
Consistent with the multiplicity and complexity of targeting signals and targeting pathways described in the previous sections, recent findings in the field of ER protein import have also extended this concept to a small assortment of ER protein translocases. Different types and arrangements of protein translocases, sometimes acting in concert, manage the translocation or insertion of a dedicated subset of incoming polypeptides (Figure 2). The different multimeric protein complexes that catalyze the insertion of an unfolded polypeptide, nascent or full-length, behave in a way very similar to targeting factors acting as a temporary safe harbor. As such, a membrane-integrated protein translocase transiently shields segments of incoming cargo and thereby facilitates the partitioning of hydrophobic TMH into the ER membrane or the translocation of soluble domains into the ER lumen. Thus, incoming polypeptides are handed over from a soluble targeting factor to a membrane-integrated translocase, both of which provide a chaperone-like environment and prevent improper interactions and the aggregation of unfolded polypeptides. Herein will summarize the central constituents of protein translocase complexes and refer the reader to the special issue on the “Mechanisms of ER Protein Import” as well as others [26][27][30][32].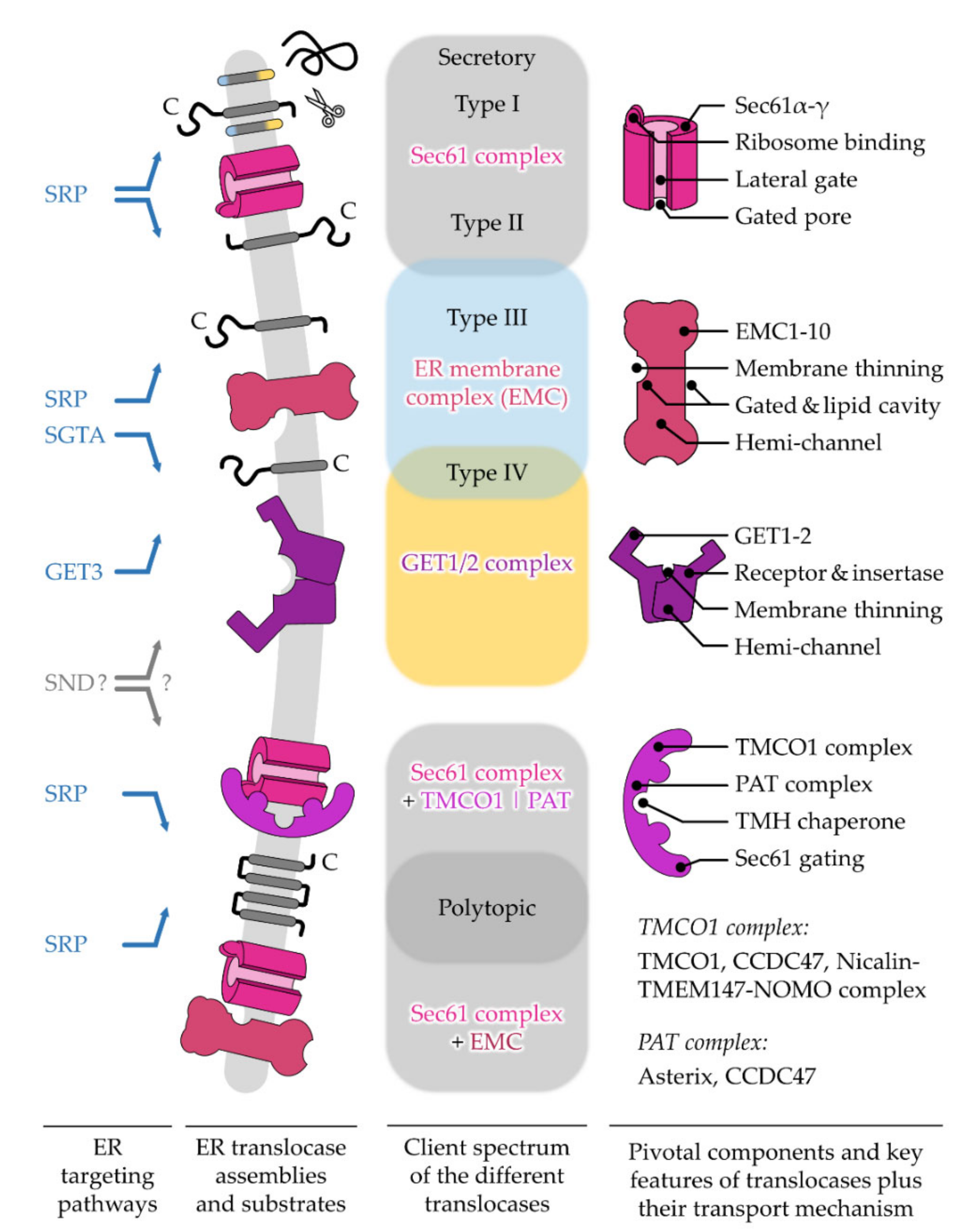
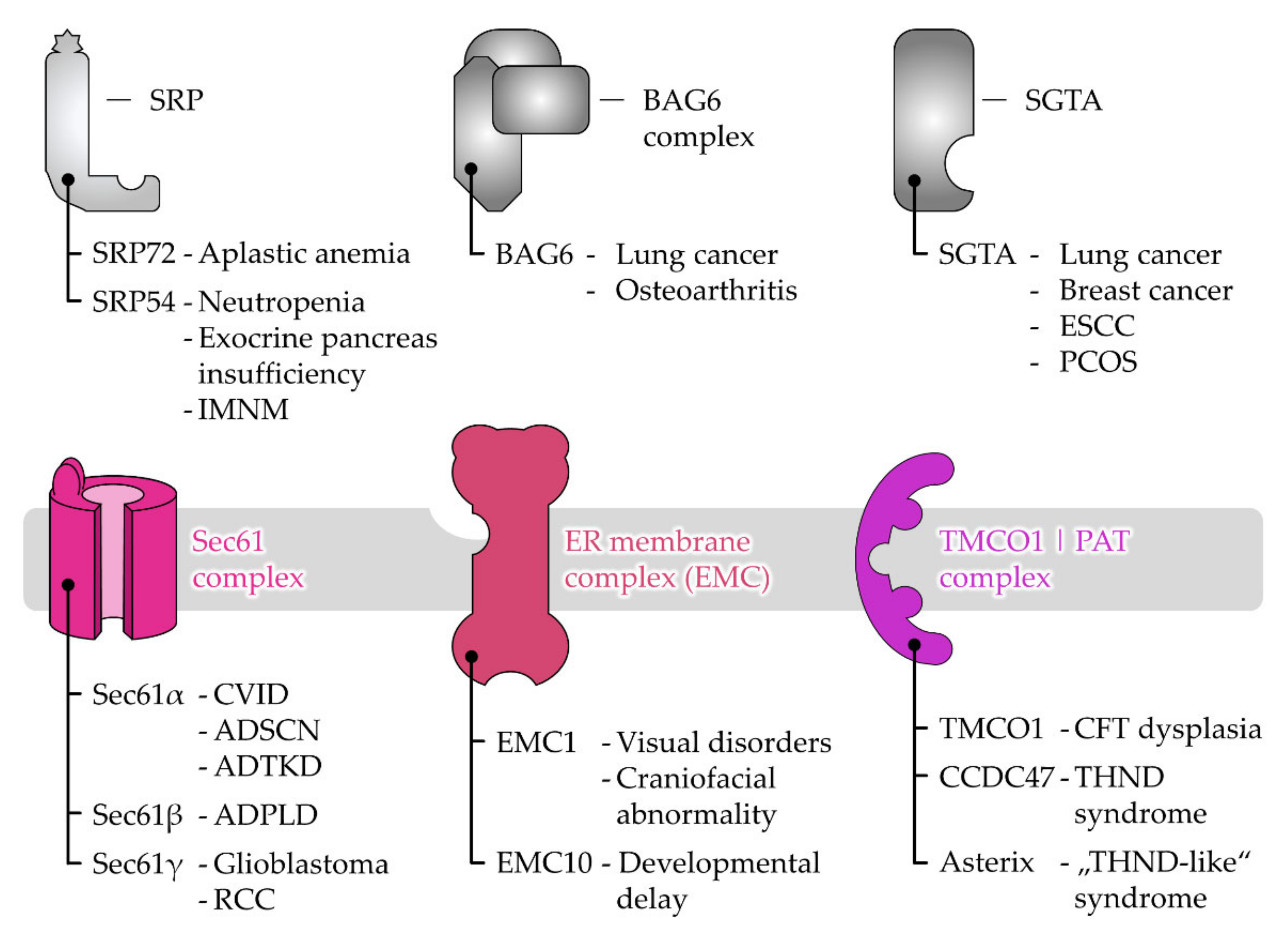
3. Disease-Causing Mutations of Targeting and Translocation Components
4. Conclusions and Perspectives
List of Abbreviations
| BAG6 | BCL2-associated athanogene 6 |
| CCDC47 | coiled-coil domain containing protein 47 |
| EMC | ER membrane protein complex |
| ER | endoplasmic reticulum |
| GET | guided entry of tail-anchored proteins |
| GPI | glycosylphosphatidylinositol |
| HSP | heat shock proteins |
| NOMO | nodal modulator protein |
| PAT | proteins associated with the ER translocon |
| RNC | Ribosome-nascent chain complex |
| SGTA | small glutamine-rich tetratricopeptide repeat-containing protein alpha |
| SND | SRP-independent targeting pathway |
| SP | signal peptide |
| SRP | signal recognition particle |
| TA | tail-anchored |
| TMCO1 | transmembrane and coiled-coil domain-containing protein 1 |
| TMEM147 | transmembrane protein 147 |
| TMH | transmembrane helix |
References
- Baum, D.A.; Baum, B. An inside-out origin for the eukaryotic cell. BMC Biol. 2014, 12, 76.
- Embley, T.M.; Martin, W. Eukaryotic evolution, changes and challenges. Nature 2006, 440, 623–630.
- Sammels, E.; Parys, J.B.; Missiaen, L.; De Smedt, H.; Bultynck, G. Intracellular Ca2+ storage in health and disease: A dynamic equilibrium. Cell Calcium 2010, 47, 297–314.
- Feske, S. Calcium signalling in lymphocyte activation and disease. Nat. Rev. Immunol. 2007, 7, 690–702.
- Valm, A.M.; Cohen, S.; Legant, W.R.; Melunis, J.; Hershberg, U.; Wait, E.; Cohen, A.R.; Davidson, M.W.; Betzig, E.; Lippincott-Schwartz, J. Applying systems-level spectral imaging and analysis to reveal the organelle interactome. Nature 2017, 546, 162–167.
- Palade, G.E. The endoplasmic reticulum. J. Biophys. Biochem. Cytol. 1956, 2, 85–98.
- Pick, T.; Beck, A.; Gamayun, I.; Schwarz, Y.; Schirra, C.; Jung, M.; Krause, E.; Niemeyer, B.A.; Zimmermann, R.; Lang, S.; et al. Remodelling of Ca2+ homeostasis is linked to enlarged endoplasmic reticulum in secretory cells. Cell Calcium 2021, 99, 102473.
- Federovitch, C.M.; Ron, D.; Hampton, R.Y. The dynamic ER: Experimental approaches and current questions. Curr. Opin. Cell Biol. 2005, 17, 409–414.
- Aviram, N.; Schuldiner, M. Targeting and translocation of proteins to the endoplasmic reticulum at a glance. J. Cell Sci. 2017, 130, 4079–4085.
- Guerriero, C.J.; Brodsky, J.L. The Delicate Balance Between Secreted Protein Folding and Endoplasmic Reticulum-Associated Degradation in Human Physiology. Physiol. Rev. 2012, 92, 537–576.
- Zimmermann, R.; Eyrisch, S.; Ahmad, M.; Helms, V. Protein translocation across the ER membrane. Biochim. Et Biophys. Acta 2011, 1808, 912–924.
- Walter, P.; Ibrahimi, I.; Blobel, G. Translocation of proteins across the endoplasmic reticulum. I. Signal recognition protein (SRP) binds to in-vitro-assembled polysomes synthesizing secretory protein. J. Cell Biol. 1981, 91, 545–550.
- Gilmore, R.; Blobel, G.; Walter, P. Protein translocation across the endoplasmic reticulum. I. Detection in the microsomal membrane of a receptor for the signal recognition particle. J. Cell Biol. 1982, 95, 463–469.
- Panzner, S.; Dreier, L.; Hartmann, E.; Kostka, S.; Rapoport, T.A. Posttranslational protein transport in yeast reconstituted with a purified complex of Sec proteins and Kar2p. Cell 1995, 81, 561–570.
- Plath, K.; Rapoport, T.A. Spontaneous Release of Cytosolic Proteins from Posttranslational Substrates before Their Transport into the Endoplasmic Reticulum. J. Cell Biol. 2000, 151, 167–178.
- Johnson, N.; Powis, K.; High, S. Post-translational translocation into the endoplasmic reticulum. Biochim. Et Biophys. Acta (BBA) Mol. Cell Res. 2013, 1833, 2403–2409.
- Haßdenteufel, S.; Nguyen, D.; Helms, V.; Lang, S.; Zimmermann, R. ER import of small human presecretory proteins: Components and mechanisms. FEBS Lett. 2019, 593, 2506–2524.
- Stefanovic, S.; Hegde, R.S. Identification of a Targeting Factor for Posttranslational Membrane Protein Insertion into the ER. Cell 2007, 128, 1147–1159.
- Schuldiner, M.; Metz, J.; Schmid, V.; Denic, V.; Rakwalska, M.; Schmitt, H.D.; Schwappach, B.; Weissman, J.S. The GET Complex Mediates Insertion of Tail-Anchored Proteins into the ER Membrane. Cell 2008, 134, 634–645.
- Johnson, N.; Vilardi, F.; Lang, S.; Leznicki, P.; Zimmermann, R.; High, S. TRC40 can deliver short secretory proteins to the Sec61 translocon. J. Cell Sci. 2012, 125, 3612–3620.
- Aviram, N.; Ast, T.; Costa, E.A.; Arakel, E.C.; Chuartzman, S.G.; Jan, C.H.; Haßdenteufel, S.; Dudek, J.; Jung, M.; Schorr, S.; et al. The SND proteins constitute an alternative targeting route to the endoplasmic reticulum. Nature 2016, 540, 134–138.
- Haßdenteufel, S.; Sicking, M.; Schorr, S.; Aviram, N.; Fecher-Trost, C.; Schuldiner, M.; Jung, M.; Zimmermann, R.; Lang, S. hSnd2 protein represents an alternative targeting factor to the endoplasmic reticulum in human cells. FEBS Lett. 2017, 591, 3211–3224.
- Casson, J.; McKenna, M.; Haßdenteufel, S.; Aviram, N.; Zimmerman, R.; High, S. Multiple pathways facilitate the biogenesis of mammalian tail-anchored proteins. J. Cell Sci. 2017, 130, 3851–3861.
- Shao, S.; Hegde, R.S. A Calmodulin-Dependent Translocation Pathway for Small Secretory Proteins. Cell 2011, 147, 1576–1588.
- Haßdenteufel, S.; Schäuble, N.; Cassella, P.; Leznicki, P.; Müller, A.; High, S.; Jung, M.; Zimmermann, R. Ca2+-calmodulin inhibits tail-anchored protein insertion into the mammalian endoplasmic reticulum membrane. FEBS Lett. 2011, 585, 3485–3490.
- Hegde, R.S.; Keenan, R.J. The mechanisms of integral membrane protein biogenesis. Nat. Rev. Mol. Cell Biol. 2021.
- O’Keefe, S.; Pool, M.R.; High, S. Membrane protein biogenesis at the ER: The highways and byways. FEBS J. 2021, n/a.
- Park, E.; Rapoport, T.A. Mechanisms of Sec61/SecY-Mediated Protein Translocation Across Membranes. Annu. Rev. Biophys. 2012, 41, 21–40.
- McDowell, M.A.; Heimes, M.; Sinning, I. Structural and molecular mechanisms for membrane protein biogenesis by the Oxa1 superfamily. Nat. Struct. Mol. Biol. 2021, 28, 234–239.
- Gemmer, M.; Förster, F. A clearer picture of the ER translocon complex. J. Cell Sci. 2020, 133, jcs231340.
- Rapoport, T.A.; Li, L.; Park, E. Structural and Mechanistic Insights into Protein Translocation. Annu. Rev. Cell Dev. Biol. 2017, 33, 369–390.
- Chitwood, P.J.; Hegde, R.S. The Role of EMC during Membrane Protein Biogenesis. Trends Cell Biol. 2019, 29, 371–384.
- Mateja, A.; Keenan, R.J. A structural perspective on tail-anchored protein biogenesis by the GET pathway. Curr. Opin. Struct. Biol. 2018, 51, 195–202.
- Blobel, G.; Sabatini, D.D. Ribosome-Membrane Interaction in Eukaryotic Cells. In Biomembranes: Volume 2, Manson, L.A., Ed.; Springer: Boston, MA, USA, 1971; pp. 193–195.
- Chitwood, P.J.; Hegde, R.S. An intramembrane chaperone complex facilitates membrane protein biogenesis. Nature 2020, 584, 630–634.
- McGilvray, P.T.; Anghel, S.A.; Sundaram, A.; Zhong, F.; Trnka, M.J.; Fuller, J.R.; Hu, H.; Burlingame, A.L.; Keenan, R.J. An ER translocon for multi-pass membrane protein biogenesis. eLife 2020, 9, e56889.
