GI microbiota dysbiosis has been associated with respiratory disorders, including COVID-19, as well as sporadic colorectal cancer (CRC) through imbalanced microbiota and compromised immune response. It is pertinent to understand the possible role of probiotics in stabilizing the microbial environment and maintaining the integrity of the respiratory and GI tracts in SARS-CoV-2 induced dysbiosis and colorectal carcinogenesis.
- gut microbiota
- colorectal cancer
- respiratory tract infection
- SARS-CoV-2
- Dysbiosis
- Virus
- Probiotic
- Lung
1. Introduction
Probiotics are defined as “live microorganisms which when administered orally in adequate amount confer a health benefit on the host” [1][2]. They are described as a live microbial feed and food supplement that beneficially affects the host’s intestinal tract [3]. Probiotics are non-pathogenic microbes that exert a variety of beneficial effects, such as antipathogenic effects, immunomodulatory factors, the production of key nutrients, and the development of mucosal epithelia. Products derived from bacteria or their end products cannot be considered probiotic because they are not alive when administered or during consumption [4]. One important point common to all these definitions is the ability of the probiotic to confer a beneficial effect on the health of the host. The implantation or colonization of these viable microorganisms improves the microbial balance of the intestinal tract. Viruses are the cause of nearly 90% of upper respiratory tract infections [5]. However, certain probiotic strains may prevent bacterial and viral diseases, such as gastroenteritis [6][7] and respiratory tract infections (RTIs), including COVID-19 [5][8][9][10][11]. It is worth noting that not all probiotics, even those that offer GI advantages, help to reduce the risk of respiratory infection in every way. For example, Lactobacillus Rhamnosus GG and Bifidobacterium animalis ssp. lactis may help the GIT, but they do not diminish the number of viruses in the nasopharynx [12]. Many in vivo and in vitro studies reveal an association between these beneficial bacteria and human immune-modulatory responses. This has led to a shift in the focus of research towards the beneficial use of probiotics in the treatment of various diseases in recent years. It is vital, therefore, to understand some of the areas regarding GIT and RTI diseases to which probiotics have been applied extensively in recent years, as well as to perform meaningful estimates for future applications, particularly in the treatment of COVID-19.
2. GI Microbiota and CRC
3. GI Microbiota Dysbiosis Associated with SARS-CoV-2 Infection and CRC
The role of the digestive system and GI microbiota in SARS-CoV-2 infection evokes the idea of the gut–lung axis. This refers to the bi-directional interplay between the GI microbiota and the lungs, which can affect immune responses and influence the course of respiratory disorders [45]. Based on previous studies and findings, a unique idea of tight connections within the microbiota-gut-lung triad as related to SARS-CoV-2 infection has emerged [46]. It is believed that the interactions within this triangle could have a direct impact on the development of COVID-19, as well as its clinical symptoms and therapy, as shown in Figure 1.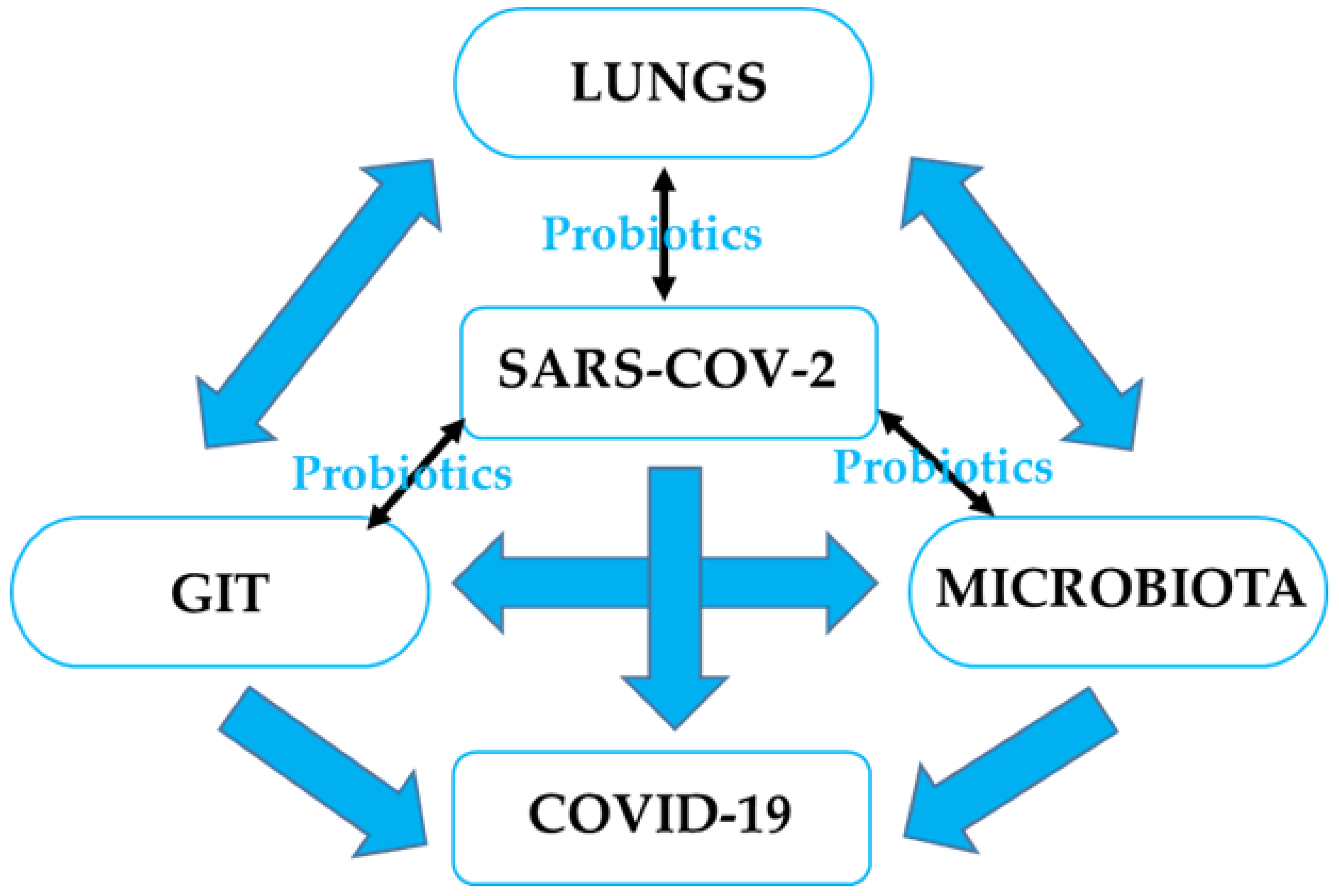
References
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S. Expert consensus document: The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514.
- Hotel, A.C.P.; Cordoba, A. Health and nutritional properties of probiotics in food including powder milk with live lactic acid bacteria. Prevention 2001, 5, 1–10.
- Vasiljevic, T.; Shah, N.P. Probiotics—from Metchnikoff to bioactive. Int. Dairy J. 2008, 18, 714–728.
- Sanders, M.E.; Gibson, G.; Gill, H.S.; Guarner, F. Probiotics: Their potential to impact human health. Counc. Agric. Sci. Technol. Issue Pap. 2007, 36, 1–20.
- Baud, D.; Dimopoulou Agri, V.; Gibson, G.R.; Reid, G.; Giannoni, E. Using probiotics to flatten the curve of coronavirus disease COVID-2019 pandemic. Front. Public Health 2020, 8, 186.
- Lenoir-Wijnkoop, I.; Gerlier, L.; Roy, D.; Reid, G. The clinical and economic impact of probiotics consumption on respiratory tract infections: Projections for Canada. PLoS ONE 2016, 11, e0166232.
- Szajewska, H.; Kołodziej, M.; Gieruszczak-Białek, D.; Skórka, A.; Ruszczyński, M.; Shamir, R. Systematic review with meta-analysis: Lactobacillus rhamnosus GG for treating acute gastroenteritis in children—A 2019 update. Aliment. Pharmacol. Ther. 2019, 49, 1376–1384.
- Guillemard, E.; Tondu, F.; Lacoin, F.; Schrezenmeir, J. Consumption of a fermented dairy product containing the probiotic Lactobacillus casei DN-114 001 reduces the duration of respiratory infections in the elderly in a randomized controlled trial. Br. J. Nutr. 2010, 103, 58–68.
- Manna, S.; Chowdhury, T.; Chakraborty, R.; Mandal, S.M. Probiotics-derived peptides and their immunomodulatory molecules can play a preventive role against viral diseases including COVID-19. Probiotics Antimicrob. Proteins 2021, 13, 611–623.
- Power, D.; Burton, J.; Chilcott, C.; Dawes, P.; Tagg, J. Preliminary investigations of the colonization of upper respiratory tract tissues of infants using a pediatric formulation of the oral probiotic Streptococcus salivarius K12. Eur. J. Clin. Microbiol. Infect. Dis. 2008, 27, 1261–1263.
- Weiss, G.; Rasmussen, S.; Zeuthen, L.H.; Nielsen, B.N.; Jarmer, H.; Jespersen, L.; Frøkiær, H. Lactobacillus acidophilus induces virus immune defense genes in murine dendritic cells by a Toll-like receptor-2-dependent mechanism. Immunology 2010, 131, 268–281.
- Lehtoranta, L.; Kalima, K.; He, L.; Lappalainen, M.; Roivainen, M.; Närkiö, M.; Mäkelä, M.; Siitonen, S.; Korpela, R.; Pitkäranta, A. Specific probiotics and virological findings in symptomatic conscripts attending military service in Finland. J. Clin. Virol. 2014, 60, 276–281.
- Hendler, R.; Zhang, Y. Probiotics in the treatment of colorectal cancer. Medicines 2018, 5, 101.
- Meng, C.; Bai, C.; Brown, T.D.; Hood, L.E.; Tian, Q. Human gut microbiota and gastrointestinal cancer. Genom. Proteom. Bioinform. 2018, 16, 33–49.
- Raskov, H.; Burcharth, J.; Pommergaard, H.-C. Linking gut microbiota to colorectal cancer. J. Cancer 2017, 8, 3378.
- Sender, R.; Fuchs, S.; Milo, R. Are we really vastly outnumbered? Revisiting the ratio of bacterial to host cells in humans. Cell 2016, 164, 337–340.
- Boleij, A.; Tjalsma, H. Gut bacteria in health and disease: A survey on the interface between intestinal microbiology and colorectal cancer. Biol. Rev. 2012, 87, 701–730.
- Aureli, P.; Capurso, L.; Castellazzi, A.M.; Clerici, M.; Giovannini, M.; Morelli, L.; Poli, A.; Pregliasco, F.; Salvini, F.; Zuccotti, G.V. Probiotics and health: An evidence-based review. Pharmacol. Res. 2011, 63, 366–376.
- Claesson, M.J.; Cusack, S.; O′Sullivan, O.; Greene-Diniz, R.; de Weerd, H.; Flannery, E.; Marchesi, J.R.; Falush, D.; Dinan, T.; Fitzgerald, G. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. S1), 4586–4591.
- Marchesi, J.R. Human distal gut microbiome. Environ. Microbiol. 2011, 13, 3088–3102.
- Hakansson, A.; Molin, G. Gut microbiota and inflammation. Nutrients 2011, 3, 637–682.
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904.
- Quigley, E.M.M. Gut microbiota and the role of probiotics in therapy. Curr. Opin. Pharmacol. 2011, 11, 593–603.
- Swidsinski, A.; Weber, J.; Loening-Baucke, V.; Hale, L.P.; Lochs, H. Spatial organization, and composition of the mucosal flora in patients with inflammatory bowel disease. J. Clin. Microbiol. 2005, 43, 3380–3389.
- Quigley, E.M.; Abu-Shanab, A. Small intestinal bacterial overgrowth. Infect. Dis. Clin. 2010, 24, 943–959.
- Srikanth, C.; McCormick, B.A. Interactions of the intestinal epithelium with the pathogen and the indigenous microbiota: A three-way crosstalk. Interdiscip. Perspect. Infect. Dis. 2008, 2008, 626827.
- Gao, Z.; Guo, B.; Gao, R.; Zhu, Q.; Qin, H. Microbiota dysbiosis is associated with colorectal cancer. Front. Microbiol. 2015, 6, 20.
- Castellarin, M.; Warren, R.L.; Freeman, J.D.; Dreolini, L.; Krzywinski, M.; Strauss, J.; Barnes, R.; Watson, P.; Allen-Vercoe, E.; Moore, R.A. Fusobacterium nucleatum infection is prevalent in human colorectal carcinoma. Genome Res. 2012, 22, 299–306.
- Marchesi, J.R.; Dutilh, B.E.; Hall, N.; Peters, W.H.; Roelofs, R.; Boleij, A.; Tjalsma, H. Towards the human colorectal cancer microbiome. PLoS ONE 2011, 6, e20447.
- Sobhani, I.; Tap, J.; Roudot-Thoraval, F.; Roperch, J.P.; Letulle, S.; Langella, P.; Corthier, G.; Van Nhieu, J.T.; Furet, J.P. Microbial dysbiosis in colorectal cancer (CRC) patients. PLoS ONE 2011, 6, e16393.
- Ray, K. Fusobacterium nucleatum found in colon cancer tissue—Could an infection cause colorectal cancer? Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 662.
- Strauss, J.; Kaplan, G.G.; Beck, P.L.; Rioux, K.; Panaccione, R.; DeVinney, R.; Lynch, T.; Allen-Vercoe, E. Invasive potential of gut mucosa-derived Fusobacterium nucleatum positively correlates with IBD status of the host. Inflamm. Bowel Dis. 2011, 17, 1971–1978.
- Flynn, K.J.; Baxter, N.T.; Schloss, P.D. Metabolic and community synergy of oral bacteria in colorectal cancer. Msphere 2016, 1, e00102–e00116.
- Kostic, A.D.; Chun, E.; Robertson, L.; Glickman, J.N.; Gallini, C.A.; Michaud, M.; Clancy, T.E.; Chung, D.C.; Lochhead, P.; Hold, G.L. Fusobacterium nucleatum potentiates intestinal tumorigenesis and modulates the tumor-immune microenvironment. Cell Host Microbe 2013, 14, 207–215.
- Hirayama, A.; Kami, K.; Sugimoto, M.; Sugawara, M.; Toki, N.; Onozuka, H.; Kinoshita, T.; Saito, N.; Ochiai, A.; Tomita, M. Quantitative metabolome profiling of colon and stomach cancer microenvironment by capillary electrophoresis time-of-flight mass spectrometry. Cancer Res. 2009, 69, 4918–4925.
- Tjalsma, H.; Boleij, A.; Marchesi, J.R.; Dutilh, B.E. A bacterial driver–passenger model for colorectal cancer: Beyond the usual suspects. Nat. Rev. Microbiol. 2012, 10, 575–582.
- Sears, C.L.; Garrett, W.S. Microbes, microbiota, and colon cancer. Cell Host Microbe 2014, 15, 317–328.
- Uronis, J.M.; Mühlbauer, M.; Herfarth, H.H.; Rubinas, T.C.; Jones, G.S.; Jobin, C. Modulation of the intestinal microbiota alters colitis-associated colorectal cancer susceptibility. PLoS ONE 2009, 4, e6026.
- Wang, X.; Allen, T.D.; May, R.J.; Lightfoot, S.; Houchen, C.W.; Huycke, M.M. Enterococcus faecalis induces aneuploidy and tetraploidy in colonic epithelial cells through a bystander effect. Cancer Res. 2008, 68, 9909–9917.
- Advani, S.M.; Advani, P.S.; Brown, D.W.; DeSantis, S.M.; Korphaisarn, K.; VonVille, H.M.; Bressler, J.; Lopez, D.S.; Davis, J.S.; Daniel, C.R. Global differences in the prevalence of the CpG island methylator phenotype of colorectal cancer. BMC Cancer 2019, 19, 964.
- Cheriyamundath, S.; Ben-Ze’ev, A. Wnt/β-Catenin target genes in colon cancer metastasis: The special case of L1cam. Cancers 2020, 12, 3444.
- McCrea, P.D.; Gottardi, C.J. Beyond β-catenin: Prospects for a larger catenin network in the nucleus. Nat. Rev. Mol. Cell Biol. 2016, 17, 55–64.
- Boland, C.R.; Goel, A. Microsatellite instability in colorectal cancer. Gastroenterology 2010, 138, 2073–2087.
- Arthur, J.C.; Gharaibeh, R.Z.; Mühlbauer, M.; Perez-Chanona, E.; Uronis, J.M.; McCafferty, J.; Fodor, A.A.; Jobin, C. Microbial genomic analysis reveals the essential role of inflammation in bacteria-induced colorectal cancer. Nat. Commun. 2014, 5, 4724.
- Enaud, R.; Prevel, R.; Ciarlo, E.; Beaufils, F.; Wieërs, G.; Guery, B.; Delhaes, L. The gut-lung axis in health and respiratory diseases: A place for inter-organ and inter-kingdom crosstalks. Front. Cell. Infect. Microbiol. 2020, 10, 9.
- Mulak, A. The impact of probiotics on interactions within the microbiota-gut-lung triad in COVID-19. Int. J. Food Sci. Nutr. 2021, 72, 577–578.
- Zuo, T.; Zhang, F.; Lui, G.C.; Yeoh, Y.K.; Li, A.Y.; Zhan, H.; Wan, Y.; Chung, A.C.; Cheung, C.P.; Chen, N. Alterations in the gut microbiota of patients with COVID-19 during the time of hospitalization. Gastroenterology 2020, 159, 944–955.
- Zuo, T.; Liu, Q.; Zhang, F.; Lui, G.C.-Y.; Tso, E.Y.; Yeoh, Y.K.; Chen, Z.; Boon, S.S.; Chan, F.K.; Chan, P.K. Depicting SARS-CoV-2 fecal viral activity in association with gut microbiota composition in patients with COVID-19. Gut 2021, 70, 276–284.
- Viana, S.D.; Nunes, S.; Reis, F. ACE2 imbalance as a key player for the poor outcomes in COVID-19 patients with age-related comorbidities–Role of gut microbiota dysbiosis. Ageing Res. Rev. 2020, 62, 101123.
- Gu, S.; Chen, Y.; Wu, Z.; Chen, Y.; Gao, H.; Lv, L.; Guo, F.; Zhang, X.; Luo, R.; Huang, C. Alterations of the gut microbiota in patients with coronavirus disease 2019 or H1N1 influenza. Clin. Infect. Dis. 2020, 71, 2669–2678.
- Zuo, T.; Zhan, H.; Zhang, F.; Liu, Q.; Tso, E.Y.; Lui, G.C.; Chen, N.; Li, A.; Lu, W.; Chan, F.K. Alterations in the fecal fungal microbiome of patients with COVID-19 during the time of hospitalization until discharge. Gastroenterology 2020, 159, 1302–1310.
- Mönkemüller, K.; Fry, L.C.; Rickes, S. Systemic inflammatory response and thrombosis due to alterations in the gut microbiota in COVID-19. Rev. Esp. Enferm. Dig. Organo Of. Soc. Esp. Patol. Dig. 2020, 112, 584–585.
- Tang, L.; Gu, S.; Gong, Y.; Li, B.; Lu, H.; Li, Q.; Zhang, R.; Gao, X.; Wu, Z.; Zhang, J. Clinical significance of the correlation between changes in the major intestinal bacteria species and COVID-19 severity. Engineering 2020, 6, 1178–1184.
- Dhar, D.; Mohanty, A. Gut microbiota and COVID-19—Possible link and implications. Virus Res. 2020, 285, 198018.
- Gou, W.; Fu, Y.; Yue, L.; Chen, G.-d.; Cai, X.; Shuai, M.; Xu, F.; Yi, X.; Chen, H.; Zhu, Y.J. Gut microbiota may underlie the predisposition of healthy individuals to COVID-19. medRxiv 2020.
- Brown, A.; Fernández, I.S.; Gordiyenko, Y.; Ramakrishnan, V. Ribosome-dependent activation of stringent control. Nature 2016, 534, 277–280.
- Brown, M.V.; Reader, J.S.; Tzima, E. Mammalian aminoacyl-tRNA synthetases: Cell signaling functions of the protein translation machinery. Vasc. Pharmacol. 2010, 52, 21–26.
- Kim, Y.; Sundrud, M.S.; Zhou, C.; Edenius, M.; Zocco, D.; Powers, K.; Zhang, M.; Mazitschek, R.; Rao, A.; Yeo, C.-Y. Aminoacyl-tRNA synthetase inhibition activates a pathway that branches from the canonical amino acid response in mammalian cells. Proc. Natl. Acad. Sci. USA 2020, 117, 8900–8911.
- Zhang, S.; Zeng, X.; Ren, M.; Mao, X.; Qiao, S. Novel metabolic and physiological functions of branched-chain amino acids: A review. J. Anim. Sci. Biotechnol. 2017, 8, 10.
- Bao, R.; Hernandez, K.; Huang, L.; Luke, J.J. ACE2 and TMPRSS2 expression by clinical, HLA, immune, and microbial correlates across 34 human cancers and matched normal tissues: Implications for SARS-CoV-2 COVID-19. J. Immunother. Cancer 2020, 8, e001020.
- Zhang, H.; Kang, Z.; Gong, H.; Xu, D.; Wang, J.; Li, Z.; Li, Z.; Cui, X.; Xiao, J.; Zhan, J. Digestive system is a potential route of COVID-19: An analysis of single-cell coexpression pattern of key proteins in the viral entry process. Gut 2020, 69, 1010–1018.
- Hoffmann, M.; Kleine-Weber, H.; Schroeder, S.; Krüger, N.; Herrler, T.; Erichsen, S.; Schiergens, T.S.; Herrler, G.; Wu, N.-H.; Nitsche, A. SARS-CoV-2 cell entry depends on ACE2 and TMPRSS2 and is blocked by a clinically proven protease inhibitor. Cell 2020, 181, 271–280.e278.
- Lamers, M.M.; Beumer, J.; van der Vaart, J.; Knoops, K.; Puschhof, J.; Breugem, T.I.; Ravelli, R.B.; Van Schayck, J.P.; Mykytyn, A.Z.; Duimel, H.Q. SARS-CoV-2 productively infects human gut enterocytes. Science 2020, 369, 50–54.
- Liu, C.; Wang, K.; Zhang, M.; Hu, X.; Hu, T.; Liu, Y.; Hu, Q.; Wu, S.; Yue, J. High expression of ACE2 and TMPRSS2 and clinical characteristics of COVID-19 in colorectal cancer patients. NPJ Precis. Oncol. 2021, 5, 1.
- Zhang, H.; Li, H.-B.; Lyu, J.-R.; Lei, X.-M.; Li, W.; Wu, G.; Lyu, J.; Dai, Z.-M. Specific ACE2 expression in small intestinal enterocytes may cause gastrointestinal symptoms and injury after 2019-nCoV infection. Int. J. Infect. Dis. 2020, 96, 19–24.
- Wu, Q.; Zhang, H.; Zhong, Y.; Chua, M.L.K.; Xie, C. Reply to colorectal cancer and COVID-19: Do we need to raise awareness and vigilance? Cancer 2021, 127, 980–981.
- Deriu, E.; Boxx, G.M.; He, X.; Pan, C.; Benavidez, S.D.; Cen, L.; Rozengurt, N.; Shi, W.; Cheng, G. Influenza virus affects intestinal microbiota and secondary Salmonella infection in the gut through type I Interferons. PLoS Pathog. 2016, 12, e1005572.
- Zha, L.; Garrett, S.; Sun, J. Salmonella infection in chronic inflammation and gastrointestinal cancer. Diseases 2019, 7, 28.
- Ma, W.-T.; Yao, X.-T.; Peng, Q.; Chen, D.-K. The protective and pathogenic roles of IL-17 in viral infections: Friend or foe? Open Biol. 2019, 9, 190109.
- Wang, X.; Ma, K.; Chen, M.; Ko, K.-H.; Zheng, B.-J.; Lu, L. IL-17A Promotes Pulmonary B-1a Cell Differentiation via Induction of Blimp-1 Expression during Influenza Virus Infection. PLoS Pathog. 2016, 12, e1005367.
- Ivanov, I.I.; de Llanos Frutos, R.; Manel, N.; Yoshinaga, K.; Rifkin, D.B.; Sartor, R.B.; Finlay, B.B.; Littman, D.R. Specific microbiota direct the differentiation of IL-17-producing T-helper cells in the mucosa of the small intestine. Cell Host. Microb. 2008, 4, 337–349.
- Mazmanian, S.K.; Round, J.L.; Kasper, D.L. A microbial symbiosis factor prevents intestinal inflammatory disease. Nature 2008, 453, 620–625.
- Sobhani, I.; Le Gouvello, S. Critical role for CD8+ FoxP3+ regulatory T cells in colon cancer immune response in humans. Gut 2009, 58, 743–744.
- Wu, S.; Rhee, K.-J.; Albesiano, E.; Rabizadeh, S.; Wu, X.; Yen, H.-R.; Huso, D.L.; Brancati, F.L.; Wick, E.; McAllister, F. A human colonic commensal promotes colon tumorigenesis via activation of T helper type 17 T cell responses. Nat. Med. 2009, 15, 1016–1022.
- van Dam, P.A.; Huizing, M.; Mestach, G.; Dierckxsens, S.; Tjalma, W.; Trinh, X.B.; Papadimitriou, K.; Altintas, S.; Vermorken, J.; Vulsteke, C.; et al. SARS-CoV-2 and cancer: Are they really partners in crime? Cancer Treat. Rev. 2020, 89, 102068.
- Xiao, F.; Tang, M.; Zheng, X.; Liu, Y.; Li, X.; Shan, H. Evidence for gastrointestinal infection of SARS-CoV-2. Gastroenterology 2020, 158, 1831–1833.e3.
- McGill, A.R.; Kahlil, R.; Dutta, R.; Green, R.; Howell, M.; Mohapatra, S.; Mohapatra, S.S. SARS-CoV-2 Immuno-pathogenesis and potential for diverse vaccines and therapies: Opportunities and challenges. Infect. Dis. Rep. 2021, 13, 102–125.
- Howell, M.C.; Green, R.; McGill, A.R.; Dutta, R.; Mohapatra, S.; Mohapatra, S.S. SARS-CoV-2-Induced Gut Microbiome Dysbiosis: Implications for Colorectal Cancer. Cancers 2021, 13, 2676.
