1. Introduction
In 2015, the average global wine production was approximately 2.8 × 10
10 L
[1,2][1][2]. The yeast
Brettanomyces bruxellensis (the anamorph
Dekkera bruxellensis) is one of the major spoilage organisms faced by the wine industry, which leads to economic losses worldwide
[3,4][3][4]. Concentrations as low as 10
3 cfu/mL of this spoilage yeast have been known to cause unpalatable off-odors and flavors
[5].
Brettanomyces spoilage is most frequently associated with red wines due, in part, to the increased use and efficiency of sulfur dioxide preservative in low pH white wines
[3].
Brettanomyces contamination has been detected in wines and wineries from all major wine producing countries around the world
[3,4][3][4]. The development of fast and effective methods for their detection and enumeration in the wine industry is therefore vital.
General plating media, including yeast extract peptone dextrose (YPD) and
Dekkera/Brettanomyces differential medium (DBDM), can be used with the addition of 20–100 mg/L of cycloheximide to detect
Dekkera/Brettanomyces bruxellensis. Due to reliability and low cost, plate counting is still the predominant method for detecting this yeast in the industry
[5,6,7][5][6][7].
B. bruxellensis typically has plate incubation times of more than 72 h (3 days) at 25–30 °C
[3,5][3][5]. Costly and time-consuming gas chromatography mass spectrometry (GC-MS) can also be used to detect these yeasts by measuring 4-ethylphenol concentrations. Real-time polymerase chain reaction (PCR) method has been shown to detect as little as 10 cells/mL in under 3 h, with highly specific detection and enumeration of targeted microorganisms
[5,7][5][7]. One disadvantage of PCR is that the wine matrix is complex, complicating DNA extraction and causing amplification problems
[8]. Microscopic examination of the distinctive morphology of
Brettanomyces is often used in conjunction with microscopic enumeration using Malassez or Thoma chambers
[8,9][8][9]. Even though specific cell functions can be determined by staining cells, this is a time consuming and highly subjective method
[8]. Malacrino et al. (2001) found that flow cytometry in combination with fluorescent dyes can be used to estimate yeast and bacteria counts in wines
[10]. This is a rapid enumeration method, with high specificity and the ability to analyze the physiological state of cells
[8].
Due to the lower growth rate of
Brettanomyces compared to other yeasts, new technologies, including impedance, could have the potential to reduce detection and enumeration times. Impedance is the effective resistance of an electrical system to alternating electric current. The impedance enumeration method measures the change in impedance caused by microbial growth in a selective medium under an alternating current (AC) field. The lower molecular size charged compounds produced during microbial growth cause the change in impedance. The BacTrac
TM system utilizes impedance splitting technology to measure the change in the impedance of the electrode (
E-value) and in the medium (
M-value). The impedance splitting technology measures the change in the impedance of the electrochemical double layer of the electrode in addition to the standard impedance signal (media impedance =
M-value). Impedance detection time is defined as the time from the start of the measurement period until the signal exceeds the specified
M- or
E-value threshold. For both methods, impedance is evaluated based on the growth of microbial cells added to the BacTrac measuring vials. Therefore, more cells result in more growth and shorter impedance detection times. A calibration curve is then used to convert the detection times to colony forming units (cfu)
[11,12,13][11][12][13].
Impedance technology has been used to enumerate
Dekkera bruxellensis, Saccharomyces cerevisiae, Candida stellata and
Hanseniaspora uvarum in a model wine,
Saccharomyces fragilis in fermented goat milk,
Bifidobacterium lactis in milk powders and
Escherichia coli in simulated milk ultra-filtrate and molluscs
[14,15,16,17,18][14][15][16][17][18]. As wine is a complex mixture of interacting chemicals, it is important to investigate the viability of impedance technology using a real wine instead of a model wine.
In thFi
s study, filter sterile red wine samples were inoculated with only
B. bruxellensis with the main objectives: (i) to investigate the relationship between plate counting and impedance detection times in red wine; and (ii) to validate and compare the ‘direct’ and ‘indirect’ impedance methods using a different red wine.
2. Validation, Advantages and Limitations of the ‘Direct’ and ‘Indirect’ Impedance
Five red wine samples of a different vintage, with concentrations ranging from 10
3 and 10
7 cfu/mL, were analyzed. The enumeration of
Brettanomyces in these samples was conducted using plate counting as a reference. Equations (2) and (3) were then used to calculate the ‘direct’ and ‘indirect’ impedance yeast concentration, respectively. A comparison between the three
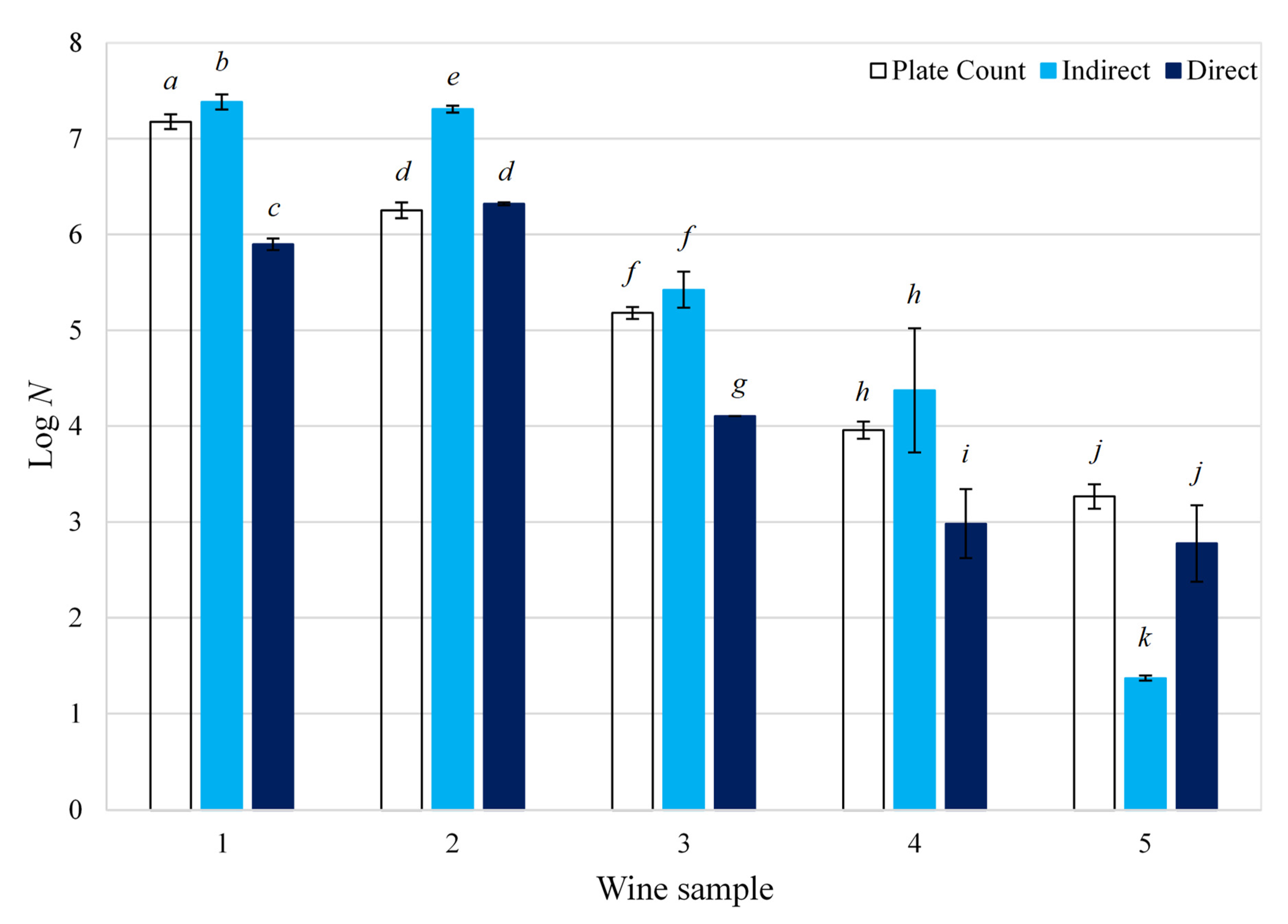
B. bruxellensis enumeration methods is presented in
Figure 1. The ‘direct’ method was slightly more accurate with an average percentage error of 15.9%, compared to that of the ‘indirect’ method, which was 18.6% (Equation (1)). For samples with a
Brettanomyces concentration above 10
4 cfu/mL, the ‘indirect’ method generally proved to be more accurate, with an average percentage error of 8.2% compared to that of the ‘direct’ method (13.2%). Contrastingly, the ‘direct’ method was more accurate at concentrations below 10
4 cfu/mL, with an average percentage error of 19.8% compared to 34.2% for the ‘indirect’ method. As expected, samples with a low yeast concentration are more difficult to quantify and thus presented results with higher variability. These results are in agreement with the results described earlier.
Figure 1. Enumeration of Brettanomyces bruxellensis contamination in wine: a comparison between plate counting and impedance technology; N is the concentration of B. bruxellensis in red wine (cfu/mL); different letters indicate that the average Log N (logarithm of yeast concentration) are significantly different according to Tukey’s test (p < 0.05).
One drawback of plate counting is the prolonged incubation periods of 120 h (5 days) required for observing
Brettanomyces colonies on the plates. Using the ‘indirect’ impedance, incubation times are reduced to 0.8 to 58 h. The reduced incubation time could be explained by the accelerated yeast proliferation rate in liquid broth compared to semi-solid agar medium, conventionally used for plate counting. Moreover, the principle of detection is different in the semi-solid agar medium contained in plates, where every cell is counted by itself, and in the liquid broth, where all cells grow together. Avoiding prolonged incubation times also decreases the chance of sample contamination during the enumeration process. Accelerated detection and enumeration times are especially important for
Brettanomyces in connection with the wine industry. This would enable faster implementation of preventative actions in the wine industry, such as the addition of more SO
2 and cleaning of equipment, to reduce the negative effects of
Brettanomyces contamination and hence wine quality degradation. The risk of large-scale wine dumping due to
Brettanomyces growth would also be reduced, in turn reducing economic losses.
One other important aspect is that standard plate counting is more labor intensive and possibly more time consuming, due to the multiple operations required for sample preparation. For the ‘direct’ and ‘indirect’ impedance methods, the sample preparation time can be significantly reduced from approximately 1 h (plate counting) to less than 5 min (impedance methods) per sample. Therefore, impedance technology has the potential to increase the throughput of enumeration analyses thereby providing faster responses to wine producers. Lastly, even though the initial capital and material costs for the impedance technology is higher, this is offset by the reduction of economic risk, labor and turnaround time of samples analyzed.
This sIt
udy i was focused on wine samples inoculated only with
B. bruxellensis. In addition, only exclusively fresh cultures of
B. bruxellensis were used in the experiments. Since the age and viability of yeasts (and other microorganisms) are different in real wines, future research on the enumeration of
B. bruxellensis in unfiltered wines should be conducted, including the identification of the impact of the age/condition of the yeast cells. Unfiltered wines contain the main fermenting organism and contaminant microorganisms (
for e
.g.xample, other yeasts and lactic acid bacteria) that can interfere with impedance detection. This could involve pre-treating the wine samples before analysis or adding antimicrobial agents to the detection media directly. Cycloheximide, to which
Brettanomyces is known to be highly resistant, has been used to isolate this yeast from real wine samples, as
S. cerevisiae’s (the predominant fermenting yeast) is susceptible to this antimicrobial additive
[21][19].
3. Conclusions
‘Direct’ and ‘indirect’ impedance were able to detect B. bruxellensis concentrations as low as 1.8 × 102 cfu/mL. The ‘indirect’ impedance has a greater potential for use in the wine industry for the enumeration of Brettanomyces, being able to decrease the processing time of wine samples containing Brettanomyces by between 62 h and 119 h (at 1.8 × 102 cfu/mL and 6.7 × 107 cfu/mL, respectively), depending on concentration. For Brettanomyces concentration above 104 cfu/mL, the ‘indirect’ method was more accurate overall compared to the ‘direct’ impedance. The ‘indirect’ impedance, in addition to offering faster detection times, is potentially less influenced by the type or composition of the wine analyzed. Therefore, the same calibration lines can possibly be used for different vintage year red wines, saving time. In conclusion, the ‘indirect’ impedance has the potential to be an alternative option for the enumeration of yeasts in the wine industry because it offers faster preparation time, high throughput and has the potential to reduce economic losses. Further research is recommended using different wines and unfiltered wines that contain other microorganisms typically present in wines.