COVIDomics, namely, the proteomic and metabolomic signatures of COVID-19. Omics-based technologies have been largely adopted during the unprecedented global COVID-19 pandemic, allowing the scientific community to perform research on a large scale to understand the pathobiology of the SARS-CoV-2 infection and its replication into human cells. The application of omics techniques has been addressed to every level of application, from the detection of mutations, methods of diagnosis or monitoring, drug target discovery, and vaccine generation, to the basic definition of the pathophysiological processes and the biochemical mechanisms behind the infection and spread of SARS-CoV-2. Thus, the term COVIDomics wants to include those efforts provided by omics-scale investigations with application to the COVID-19 research.
- COVID-19
- SARS-CoV-2
- COVIDomics
- proteomics
- metabolomics
- multiomics
- COVID-19 signature
- data integration
- pandemic
1. Introduction
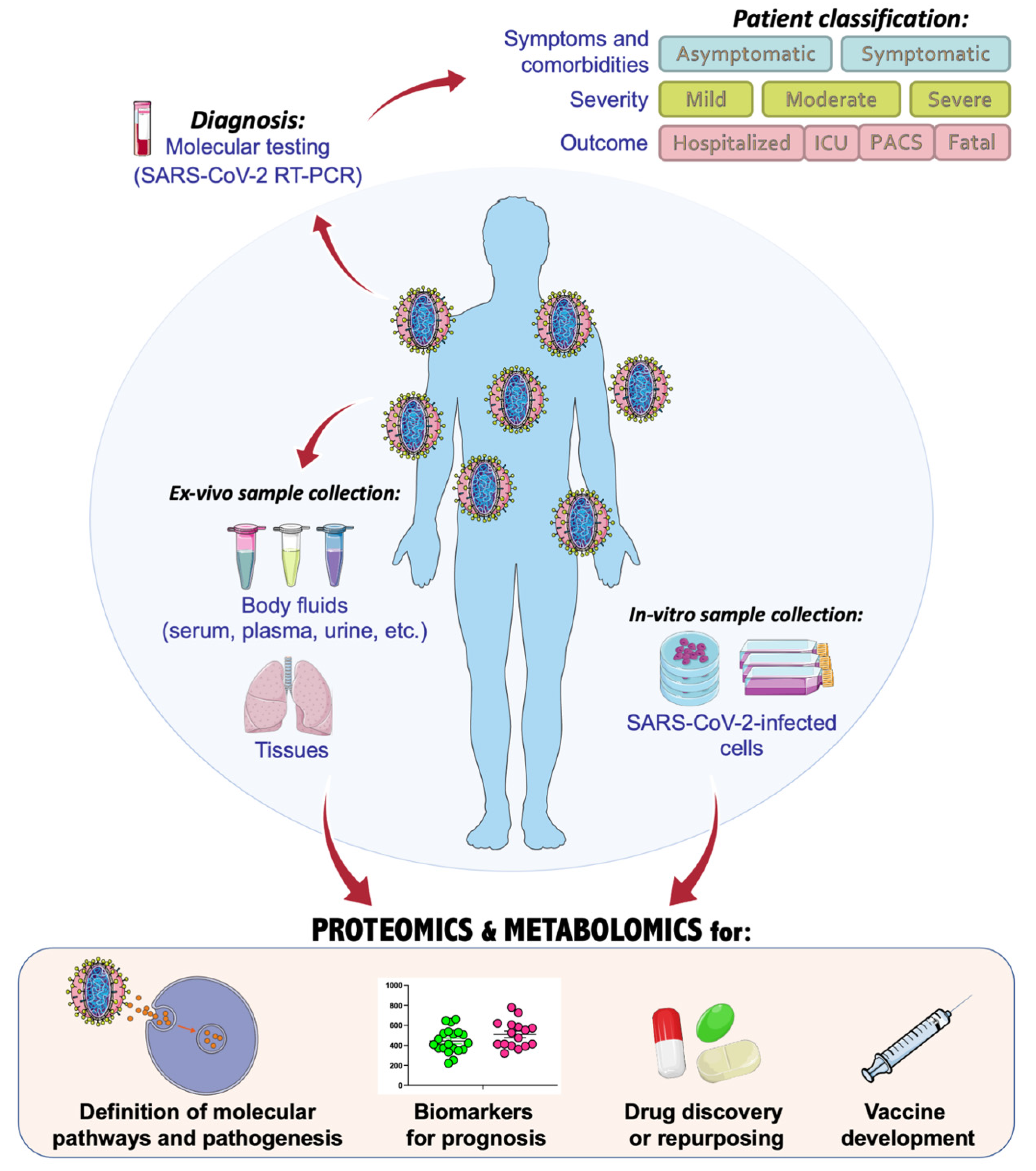
2. COVIDomics Data Integration
The application of omics and multiomics technologies, enclosed here under the term COVIDomics, offers the great chance to meticulously recognize and dissect the multiple molecular aspects that characterize a complex disease such as COVID-19. The majority of patients that contract the SARS-CoV-2 infection may develop no or mild symptoms, although many others experience a severe or critical symptomatology leading to fatal cases. This high heterogeneity in the clinical manifestation is being investigated in many studies, comprising those here concluded. In fact, many comparisons investigate the differences between the severe disease toward the baseline proteome or metabolome in healthy patients; others focus mostly on the progression of the disease from a condition to another one and highlight correlations of clinical and biochemical parameters with dysregulated proteins and metabolites. Furthermore, the majority of the COVIDomics research was performed on blood, as it is an easily accessible liquid biopsy reflecting the systemic changes subsequent the strong infection of SARS-CoV-2. Thus, the comprehensive and accurate analysis of circulating molecules is helpful to unravel the systemic mechanisms of the disease. In fact, as summarized in Figure 2 and detailed in Table 4, there are several common proteins that were identified in plasma and serum as quantitatively changed between the patients and their controls, representing a relevant proteomic signature of COVID-19.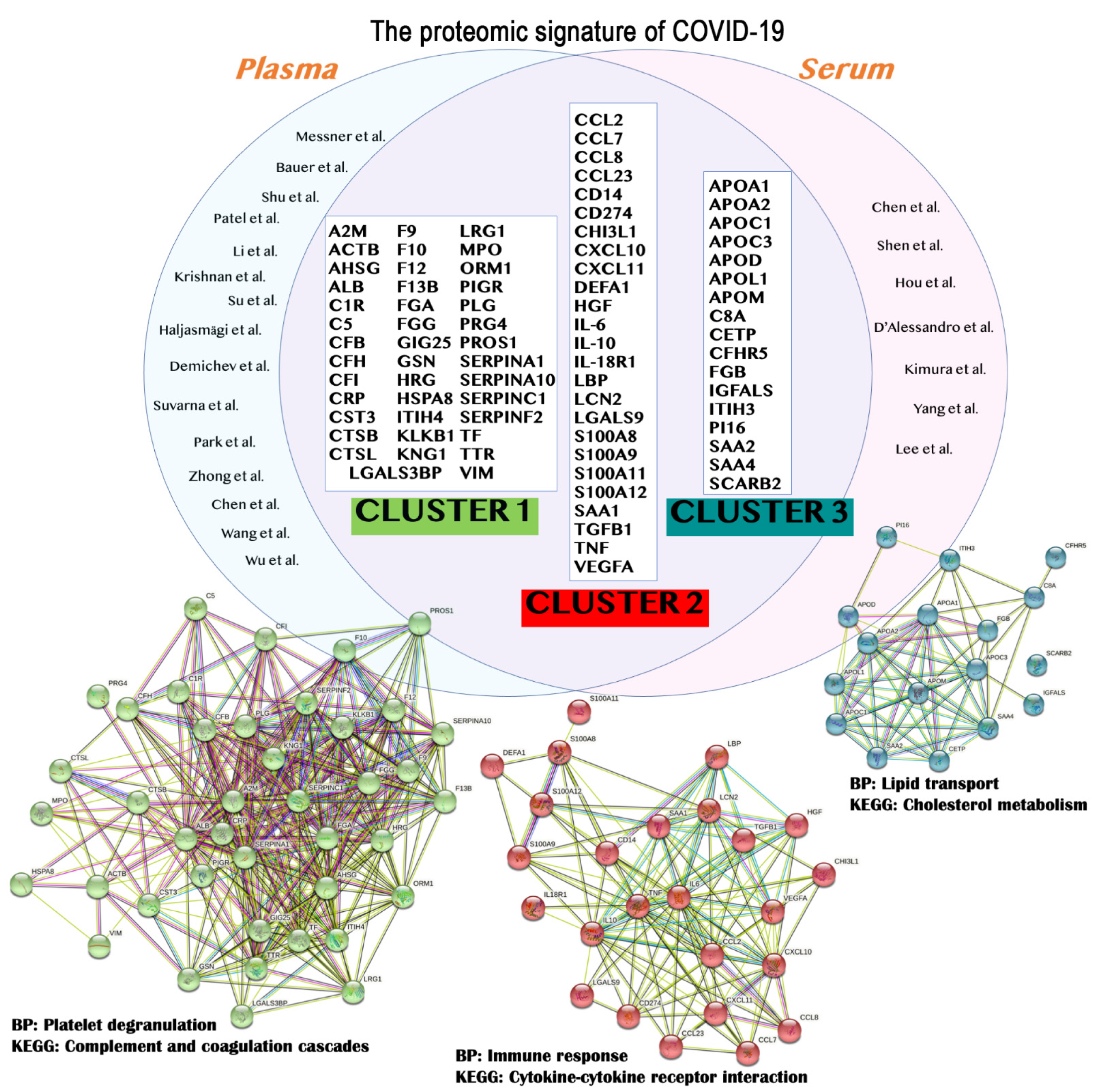
| Protein Symbol | UniProt ID | Protein Name | STRING Cluster |
|---|---|---|---|
| A2M | P01023 | Alpha-2-macroglobulin | Cluster 1 |
| ACTB | P60709 | Actin, cytoplasmic 1 | |
| AHSG | P02765 | Alpha-2-HS-glycoprotein | |
| ALB | P02768 | Albumin | |
| C1R | P00736 | Complement C1r subcomponent | |
| C5 | P01031 | Complement C5 | |
| CFB | P00751 | Complement Factor B | |
| CFH | P08603 | Complement factor H | |
| CFI | P05156 | Complement factor I | |
| CRP | P02741 | C-reactive protein | |
| CST3 | P01034 | Cystatin-C | |
| CTSB | P07858 | Cathepsin B | |
| CTSL | P07711 | Procathepsin L | |
| F9 | P00740 | Coagulation factor IX | |
| F10 | P00742 | Coagulation factor X | |
| F12 | P00748 | Coagulation factor XII | |
| F13B | P05160 | Coagulation factor XIII B chain | |
| FGA | P02671 | Fibrinogen alpha chain | |
| FGG | P02679 | Fibrinogen gamma chain | |
| GSN | P06396 | Gelsolin | |
| HRG | P04196 | Histidine-rich glycoprotein | |
| HSPA8 | P11142 | Heat shock cognate 71 kDa protein | |
| ITIH4 | Q14624 | Inter-alpha-trypsin inhibitor heavy chain H4 | |
| KLKB1 | P03952 | Plasma kallikrein | |
| KNG1 | P01042 | Kininogen-1 | |
| LGALS3BP | Q08380 | Galectin-3-binding protein | |
| LRG1 | P02750 | Leucine-rich alpha-2-glycoprotein | |
| MPO | P05164 | Myeloperoxidase | |
| ORM1 | P02763 | Alpha-1-acid glycoprotein 1 | |
| PIGR | P01833 | Polymeric immunoglobulin receptor | |
| PLG | P00747 | Plasminogen | |
| PRG4 | Q92954 | Proteoglycan 4 | |
| PROS1 | P07225 | Vitamin K-dependent protein S | |
| SERPINA1 | P01009 | Alpha-1-antitrypsin | |
| SERPINA3 | P01011 | Alpha-1-antichymotrypsin | |
| SERPINA10 | Q9UK55 | Protein Z-dependent protease inhibitor | |
| SERPINC1 | P01008 | Antithrombin-III | |
| SERPINF2 | P08697 | Alpha-2-antiplasmin | |
| TF | P02787 | Transferrin | |
| TTR | P02766 | Transthyretin | |
| VIM | P08670 | Vimentin | |
| CCL2 | P13500 | C-C motif chemokine 2 | Cluster 2 |
| CCL7 | P80098 | C-C motif chemokine 7 | |
| CCL8 | P80075 | C-C motif chemokine 8 | |
| CD14 | P08571 | Monocyte differentiation antigen CD14 | |
| CCL23 | P55773 | C-C motif chemokine 23 | |
| CD274 | Q9NZQ7 | Programmed cell death 1 ligand 1 | |
| CHI3L1 | P36222 | Chitinase-3-like protein 1 | |
| CXCL10 | P02778 | C-X-C motif chemokine 10 | |
| CXCL11 | O14625 | C-X-C motif chemokine 11 | |
| DEFA1 | P59665 | Neutrophil defensin 1 | |
| HGF | P14210 | Hepatocyte growth factor | |
| IL-10 | P22301 | Interleukin-10 | |
| IL-18R1 | Q13478 | Interleukin-18 receptor 1 | |
| IL-6 | P08887 | Interleukin-6 receptor subunit alpha | |
| LBP | P18428 | Lipopolysaccharide-binding protein | |
| LCN2 | P80188 | Neutrophil gelatinase-associated lipocalin | |
| LGALS9 | O00182 | Galectin-9 | |
| S100A11 | P31949 | Protein S100-A11 | |
| S100A12 | P80511 | Protein S100-A12 | |
| S100A8 | P05109 | Protein S100-A8 | |
| S100A9 | P06702 | Protein S100-A9 | |
| SAA1 | P0DJI8 | Serum amyloid A-1 protein | |
| TGFB1 | P01137 | Transforming growth factor beta-1 proprotein | |
| TNF | P01375 | Tumor necrosis factor | |
| VEGFA | P15692 | Vascular endothelial growth factor A | |
| APOA1 | P02647 | Apolipoprotein A1 | Cluster 3 |
| APOA2 | P02652 | Apolipoprotein A2 | |
| APOC1 | P02654 | Apolipoprotein C1 | |
| APOC3 | P02656 | Apolipoprotein C3 | |
| APOD | P05090 | Apolipoprotein D | |
| APOL1 | O14791 | Apolipoprotein L1 | |
| APOM | O95445 | Apolipoprotein M | |
| C8A | P07357 | Complement component C8 alpha chain | |
| CETP | P11597 | Cholesteryl ester transfer protein | |
| CFHR5 | Q9BXR6 | Complement factor H-related protein 5 | |
| FGB | P02675 | Fibrinogen beta chain | |
| IGFALS | P35858 | Insulin-like growth factor-binding protein complex acid labile subunit | |
| ITIH3 | Q06033 | Inter-alpha-trypsin inhibitor heavy chain H3 | |
| PI16 | Q6UXB8 | Peptidase inhibitor 16 | |
| SAA2 | P0DJI9 | Serum amyloid A-2 protein | |
| SAA4 | P35542 | Serum amyloid A-4 protein | |
| SCARB2 | Q14108 | Lysosome membrane protein 2 |
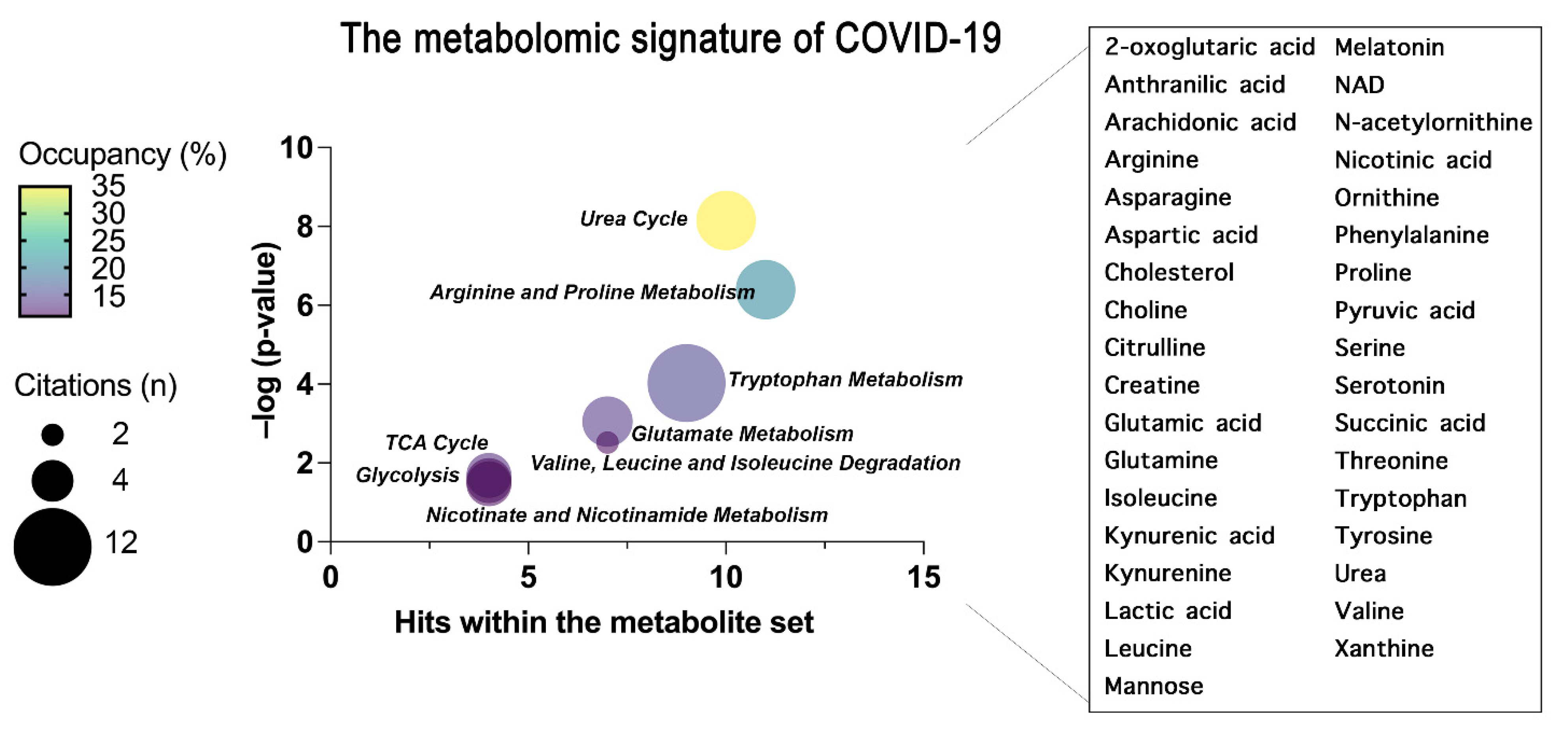 Figure 3. The common metabolites and metabolic pathways obtained from the conclusion of plasma and serum metabolomics studies are summarized. A specific signature of the metabolome of COVID-19 patients was obtained representing the most enriched pathways constructed through an over-representation analysis (ORA) within MetaboAnalyst 5.0 and represented as bubble plot. The bubble plot takes into account the statistical significance (–log p-value) of each metabolite set identified through the ORA analysis. Where the size of each bubble refers to the number of times the metabolite set is cited here by each scientist, the color instead refers to the number of metabolites detected over the total number of metabolites within that pathway (occupancy). On the right, the metabolites common to all the concluded papers that have generated the pathway enrichment are grouped.
Figure 3. The common metabolites and metabolic pathways obtained from the conclusion of plasma and serum metabolomics studies are summarized. A specific signature of the metabolome of COVID-19 patients was obtained representing the most enriched pathways constructed through an over-representation analysis (ORA) within MetaboAnalyst 5.0 and represented as bubble plot. The bubble plot takes into account the statistical significance (–log p-value) of each metabolite set identified through the ORA analysis. Where the size of each bubble refers to the number of times the metabolite set is cited here by each scientist, the color instead refers to the number of metabolites detected over the total number of metabolites within that pathway (occupancy). On the right, the metabolites common to all the concluded papers that have generated the pathway enrichment are grouped.
3. Concluding Remarks
During the current COVID-19 pandemic, unprecedented efforts have been made by the scientific community to dissect the molecular bases of SARS-CoV-2 infection, spreading, and pathogenicity. Omics scientists deserved particular merits for performing several proteomics- and metabolomics-based investigations, but also integrative multiomics analyses, using samples derived from patients. Thus, the application of COVIDomics strategies has certainly empowered the knowledge relative to COVID-19 in terms of biomarkers of disease and specific physiological mechanisms disturbed upon SARS-CoV-2 infection. The study of coronavirus structure, replication, and infection, together with the virus-host interaction and the host response, have remarkably provided us with efficient tools to diagnose the disease and ameliorate the fate of COVID-19 patients. In general, a huge improvement in patients’ outcomes is correlated to the development of vaccines as prophylactic therapeutic approaches, despite it remaining not currently possible to foresee the prognosis of infected patients by a single analysis or through the detection of a single and specific marker. Nonetheless, in this scenario, the use of omics sciences and their integration have provided us with relevant proteomics and metabolomics signatures, as a point of junction between SARS-CoV-2 infection/pathobiology with the subsequent clinical outcome of affected patients. Thus, COVIDomics is contributing to a better comprehension of the molecular intricacy of COVID-19, in the view of identifying new molecular actors involved in the pathogenesis of the disease to be used for efficient therapeutic approaches in combatting the COVID-19 pandemic.References
- Costanzo, M.; De Giglio, M.A.R.; Roviello, G.N. Anti-Coronavirus Vaccines: Past Investigations on SARS-CoV-1 and MERS-CoV, the Approved Vaccines from BioNTech/Pfizer, Moderna, Oxford/AstraZeneca and others under Development Against SARS-CoV- 2 Infection. Curr. Med. Chem. 2022, 29, 4–18.
- De Giglio, M.A.R.; Roviello, G.N. SARS-CoV-2: Recent Reports on Antiviral Therapies Based on Lopinavir/Ritonavir, Darunavir/Umifenovir, Hydroxychloroquine, Remdesivir, Favipiravir and other Drugs for the Treatment of the New Coronavirus. Curr. Med. Chem. 2020, 27, 4536–4541.
- Rehman, S.U.; Rehman, S.U.; Yoo, H.H. COVID-19 challenges and its therapeutics. Biomed. Pharmacother. 2021, 142, 112015.
- Singh, T.U.; Parida, S.; Lingaraju, M.C.; Kesavan, M.; Kumar, D.; Singh, R.K. Drug repurposing approach to fight COVID-19. Pharmacol. Rep. 2020, 72, 1479–1508.
- Borbone, N.; Piccialli, G.; Roviello, G.N.; Oliviero, G. Nucleoside Analogs and Nucleoside Precursors as Drugs in the Fight against SARS-CoV-2 and Other Coronaviruses. Molecules 2021, 26, 986.
- Xu, Z.; Shi, L.; Wang, Y.; Zhang, J.; Huang, L.; Zhang, C.; Liu, S.; Zhao, P.; Liu, H.; Zhu, L.; et al. Pathological findings of COVID-19 associated with acute respiratory distress syndrome. Lancet Respir. Med. 2020, 8, 420–422.
- Nalbandian, A.; Sehgal, K.; Gupta, A.; Madhavan, M.V.; McGroder, C.; Stevens, J.S.; Cook, J.R.; Nordvig, A.S.; Shalev, D.; Sehrawat, T.S.; et al. Post-acute COVID-19 syndrome. Nat. Med. 2021, 27, 601–615.
- Marshall, J.C.; Murthy, S.; Diaz, J.; Adhikari, N.K.; Angus, D.C.; Arabi, Y.M.; Baillie, K.; Bauer, M.; Berry, S.; Blackwood, B.; et al. A minimal common outcome measure set for COVID-19 clinical research. Lancet Infect. Dis. 2020, 20, e192–e197.
- Zhang, Y.-Z.; Holmes, E.C. A Genomic Perspective on the Origin and Emergence of SARS-CoV-2. Cell 2020, 181, 223–227.
- Naqvi, A.A.T.; Fatima, K.; Mohammad, T.; Fatima, U.; Singh, I.K.; Singh, A.; Atif, S.M.; Hariprasad, G.; Hasan, G.M.; Hassan, I. Insights into SARS-CoV-2 genome, structure, evolution, pathogenesis and therapies: Structural genomics approach. Biochim. Biophys. Acta (BBA) Mol. Basis Dis. 2020, 1866, 165878.
- Bell, L.C.; Meydan, C.; Kim, J.; Foox, J.; Butler, D.; Mason, C.E.; Shapira, S.D.; Noursadeghi, M.; Pollara, G. Transcriptional response modules characterize IL-1β and IL-6 activity in COVID-19. iScience 2021, 24, 101896.
- Fagone, P.; Ciurleo, R.; Lombardo, S.D.; Iacobello, C.; Palermo, C.I.; Shoenfeld, Y.; Bendtzen, K.; Bramanti, P.; Nicoletti, F. Transcriptional landscape of SARS-CoV-2 infection dismantles pathogenic pathways activated by the virus, proposes unique sex-specific differences and predicts tailored therapeutic strategies. Autoimmun. Rev. 2020, 19, 102571.
- Cho, K.-C.; Clark, D.J.; Schnaubelt, M.; Teo, G.C.; Leprevost, F.D.V.; Bocik, W.; Boja, E.S.; Hiltke, T.; Nesvizhskii, A.I.; Zhang, H. Deep Proteomics Using Two Dimensional Data Independent Acquisition Mass Spectrometry. Anal. Chem. 2020, 92, 4217–4225.
- Kawashima, Y.; Watanabe, E.; Umeyama, T.; Nakajima, D.; Hattori, M.; Honda, K.; Ohara, O. Optimization of Data-Independent Acquisition Mass Spectrometry for Deep and Highly Sensitive Proteomic Analysis. Int. J. Mol. Sci. 2019, 20, 5932.
- Nakajima, D.; Ohara, O.; Kawashima, Y. Data-Independent Acquisition Mass Spectrometry-Based Deep Proteome Analysis for Hydrophobic Proteins from Dried Blood Spots Enriched by Sodium Carbonate Precipitation. In Clinical Proteomics; Humana: New Yor, NY, USA, 2022; pp. 39–52. ISBN 978-1-0716-1935-3.
- Costanzo, M.; Caterino, M.; Cevenini, A.; Kollipara, L.; Shevchuk, O.; Nguyen, C.D.L.; Sickmann, A.; Ruoppolo, M. Data Independent Acquisition Mass Spectrometry for Proteomic Advances into Isolated Methylmalonic Acidemia. In NATO Science for Peace and Security Series A: Chemistry and Biology; Springer: Dordrecht, The Netherlands, 2020; pp. 221–223.
- Barigazzi, E.; Santorelli, L.; Morello, W.; Raimondo, F.; Crapella, B.; Ghio, L.; Tamburello, C.; Montini, G.; Pitto, M. New Insight into Idiopathic Nephrotic Syndrome: Strategy Based on Urinary Exosomes. In NATO Science for Peace and Security Series A: Chemistry and Biology; Springer: Dordrecht, The Netherlands, 2020; pp. 217–218.
- Santorelli, L.; Stella, M.; Chinello, C.; Capitoli, G.; Piga, I.; Smith, A.; Grasso, A.; Grasso, M.; Bovo, G.; Magni, F. Does the Urinary Proteome Reflect ccRCC Stage and Grade Progression? Diagnostics 2021, 11, 2369.
- Kong, S.W.; Hernandez-Ferrer, C. Assessment of coverage for endogenous metabolites and exogenous chemical compounds using an untargeted metabolomics platform. Pac. Symp. Biocomput. 2020, 25, 587–598.
- Manganelli, V.; Salvatori, I.; Costanzo, M.; Capozzi, A.; Caissutti, D.; Caterino, M.; Valle, C.; Ferri, A.; Sorice, M.; Ruoppolo, M.; et al. Overexpression of Neuroglobin Promotes Energy Metabolism and Autophagy Induction in Human Neuroblastoma SH-SY5Y Cells. Cells 2021, 10, 3394.
- Costanzo, M.; Caterino, M.; Cevenini, A.; Jung, V.; Chhuon, C.; Lipecka, J.; Fedele, R.; Guerrera, I.C.; Ruoppolo, M. Dataset of a comparative proteomics experiment in a methylmalonyl-CoA mutase knockout HEK 293 cell model. Data Brief 2020, 33, 106453.
- Gordon, D.E.; Jang, G.M.; Bouhaddou, M.; Xu, J.; Obernier, K.; White, K.M.; O’Meara, M.J.; Rezelj, V.V.; Guo, J.Z.; Swaney, D.L.; et al. A SARS-CoV-2 protein interaction map reveals targets for drug repurposing. Nature 2020, 583, 459–468.
- Giacco, A.; Paoli, G.D.; Senese, R.; Cioffi, F.; Silvestri, E.; Moreno, M.; Ruoppolo, M.; Caterino, M.; Costanzo, M.; Lombardi, A.; et al. The saturation degree of fatty acids and their derived acylcarnitines determines the direct effect of metabolically active thyroid hormones on insulin sensitivity in skeletal muscle cells. FASEB J. 2018, 33, 1811–1823.
- Ruoppolo, M.; Caterino, M.; Albano, L.; Pecce, R.; Di Girolamo, M.G.; Crisci, D.; Costanzo, M.; Milella, L.; Franconi, F.; Campesi, I. Targeted metabolomic profiling in rat tissues reveals sex differences. Sci. Rep. 2018, 8, 4663.
- De Pasquale, V.; Caterino, M.; Costanzo, M.; Fedele, R.; Ruoppolo, M.; Pavone, L.M. Targeted Metabolomic Analysis of a Mucopolysaccharidosis IIIB Mouse Model Reveals an Imbalance of Branched-Chain Amino Acid and Fatty Acid Metabolism. Int. J. Mol. Sci. 2020, 21, 4211.
- Heiles, S. Advanced tandem mass spectrometry in metabolomics and lipidomics—methods and applications. Anal. Bioanal. Chem. 2021, 413, 5927–5948.
- Rampler, E.; El Abiead, Y.; Schoeny, H.; Rusz, M.; Hildebrand, F.; Fitz, V.; Koellensperger, G. Recurrent Topics in Mass Spectrometry-Based Metabolomics and Lipidomics—Standardization, Coverage, and Throughput. Anal. Chem. 2020, 93, 519–545.
- Abu-Farha, M.; Thanaraj, T.A.; Qaddoumi, M.G.; Hashem, A.; Abubaker, J.; Al-Mulla, F. The Role of Lipid Metabolism in COVID-19 Virus Infection and as a Drug Target. Int. J. Mol. Sci. 2020, 21, 3544.
- Galbraith, M.D.; Kinning, K.T.; Sullivan, K.D.; Baxter, R.; Araya, P.; Jordan, K.R.; Russell, S.; Smith, K.P.; Granrath, R.E.; Shaw, J.R.; et al. Seroconversion stages COVID19 into distinct pathophysiological states. eLife 2021, 10.
- Li, C.-X.; Gao, J.; Zhang, Z.; Chen, L.; Li, X.; Zhou, M.; Wheelock, Å.M. Multiomics integration-based molecular characterizations of COVID-19. Brief. Bioinform. 2021, 23, bbab485.
- Patel, H.; Ashton, N.J.; Dobson, R.J.B.; Andersson, L.-M.; Yilmaz, A.; Blennow, K.; Gisslen, M.; Zetterberg, H. Proteomic blood profiling in mild, severe and critical COVID-19 patients. Sci. Rep. 2021, 11, 6357.
- Haljasmägi, L.; Salumets, A.; Rumm, A.P.; Jürgenson, M.; Krassohhina, E.; Remm, A.; Sein, H.; Kareinen, L.; Vapalahti, O.; Sironen, T.; et al. Longitudinal proteomic profiling reveals increased early inflammation and sustained apoptosis proteins in severe COVID-19. Sci. Rep. 2020, 10, 20533.
- Zhong, W.; Altay, O.; Arif, M.; Edfors, F.; Doganay, L.; Mardinoglu, A.; Uhlen, M.; Fagerberg, L. Next generation plasma proteome profiling of COVID-19 patients with mild to moderate symptoms. eBioMedicine 2021, 74, 103723.
- Bauer, W.; Weber, M.; Diehl-Wiesenecker, E.; Galtung, N.; Prpic, M.; Somasundaram, R.; Tauber, R.; Schwenk, J.M.; Micke, P.; Kappert, K. Plasma Proteome Fingerprints Reveal Distinctiveness and Clinical Outcome of SARS-CoV-2 Infection. Viruses 2021, 13, 2456.
- Shu, T.; Ning, W.; Wu, D.; Xu, J.; Han, Q.; Huang, M.; Zou, X.; Yang, Q.; Yuan, Y.; Bie, Y.; et al. Plasma Proteomics Identify Biomarkers and Pathogenesis of COVID-19. Immunity 2020, 53, 1108–1122.e5.
- Park, J.; Kim, H.; Kim, S.Y.; Kim, Y.; Lee, J.-S.; Dan, K.; Seong, M.-W.; Han, D. In-depth blood proteome profiling analysis revealed distinct functional characteristics of plasma proteins between severe and non-severe COVID-19 patients. Sci. Rep. 2020, 10, 22418.
- Messner, C.B.; Demichev, V.; Wendisch, D.; Michalick, L.; White, M.; Freiwald, A.; Textoris-Taube, K.; Vernardis, S.I.; Egger, A.-S.; Kreidl, M.; et al. Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of COVID-19 Infection. Cell Syst. 2020, 11, 11–24.
- Demichev, V.; Tober-Lau, P.; Lemke, O.; Nazarenko, T.; Thibeault, C.; Whitwell, H.; Röhl, A.; Freiwald, A.; Szyrwiel, L.; Ludwig, D.; et al. A time-resolved proteomic and prognostic map of COVID-19. Cell Syst. 2021, 12, 780–794.e7.
- Su, Y.; Chen, D.; Yuan, D.; Lausted, C.; Choi, J.; Dai, C.L.; Voillet, V.; Duvvuri, V.R.; Scherler, K.; Troisch, P.; et al. Multi-Omics Resolves a Sharp Disease-State Shift between Mild and Moderate COVID-19. Cell 2020, 183, 1479–1495.e20.
- Chen, Y.; Zheng, Y.; Yu, Y.; Wang, Y.; Huang, Q.; Qian, F.; Sun, L.; Song, Z.; Chen, Z.; Feng, J.; et al. Blood molecular markers associated with COVID-19 immunopathology and multi-organ damage. EMBO J. 2020, 39, e105896.
- Krishnan, S.; Nordqvist, H.; Ambikan, A.T.; Gupta, S.; Sperk, M.; Svensson-Akusjärvi, S.; Mikaeloff, F.; Benfeitas, R.; Saccon, E.; Ponnan, S.M.; et al. Metabolic Perturbation Associated With COVID-19 Disease Severity and SARS-CoV-2 Replication. Mol. Cell. Proteom. 2021, 20, 100159.
- Suvarna, K.; Salkar, A.; Palanivel, V.; Bankar, R.; Banerjee, N.; Pai, M.G.J.; Srivastava, A.; Singh, A.; Khatri, H.; Agrawal, S.; et al. A Multi-omics Longitudinal Study Reveals Alteration of the Leukocyte Activation Pathway in COVID-19 Patients. J. Proteome Res. 2021, 20, 4667–4680.
- Wu, P.; Chen, D.; Ding, W.; Wu, P.; Hou, H.; Bai, Y.; Zhou, Y.; Li, K.; Xiang, S.; Liu, P.; et al. The trans-omics landscape of COVID-19. Nat. Commun. 2021, 12, 4543.
- Li, Y.; Hou, G.; Zhou, H.; Wang, Y.; Tun, H.M.; Zhu, A.; Zhao, J.; Xiao, F.; Lin, S.; Liu, D.; et al. Multi-platform omics analysis reveals molecular signature for COVID-19 pathogenesis, prognosis and drug target discovery. Signal Transduct. Target. Ther. 2021, 6, 155.
- Wang, C.; Li, X.; Ning, W.; Gong, S.; Yang, F.; Fang, C.; Gong, Y.; Wu, D.; Huang, M.; Gou, Y.; et al. Multi-omic profiling of plasma reveals molecular alterations in children with COVID-19. Theranostics 2021, 11, 8008–8026.
- Hou, X.; Zhang, X.; Wu, X.; Lu, M.; Wang, D.; Xu, M.; Wang, H.; Liang, T.; Dai, J.; Duan, H.; et al. Serum Protein Profiling Reveals a Landscape of Inflammation and Immune Signaling in Early-stage COVID-19 Infection. Mol. Cell. Proteom. 2020, 19, 1749–1759.
- Chen, Y.; Yao, H.; Zhang, N.; Wu, J.; Gao, S.; Guo, J.; Lu, X.; Cheng, L.; Luo, R.; Liang, X.; et al. Proteomic Analysis Identifies Prolonged Disturbances in Pathways Related to Cholesterol Metabolism and Myocardium Function in the COVID-19 Recovery Stage. J. Proteome Res. 2021, 20, 3463–3474.
- Kimura, Y.; Nakai, Y.; Shin, J.; Hara, M.; Takeda, Y.; Kubo, S.; Jeremiah, S.S.; Ino, Y.; Akiyama, T.; Moriyama, K.; et al. Identification of serum prognostic biomarkers of severe COVID-19 using a quantitative proteomic approach. Sci. Rep. 2021, 11, 20638.
- Lee, J.; Han, D.; Kim, S.Y.; Hong, K.H.; Jang, M.; Kim, M.J.; Kim, Y.; Park, J.H.; Cho, S.I.; Park, W.B.; et al. Longitudinal proteomic profiling provides insights into host response and proteome dynamics in COVID-19 progression. Proteomics 2021, 21, 2000278.
- D’Alessandro, A.; Thomas, T.; Dzieciatkowska, M.; Hill, R.C.; Francis, R.O.; Hudson, K.E.; Zimring, J.C.; Hod, E.A.; Spitalnik, S.L.; Hansen, K.C. Serum Proteomics in COVID-19 Patients: Altered Coagulation and Complement Status as a Function of IL-6 Level. J. Proteome Res. 2020, 19, 4417–4427.
- Shen, B.; Yi, X.; Sun, Y.; Bi, X.; Du, J.; Zhang, C.; Quan, S.; Zhang, F.; Sun, R.; Qian, L.; et al. Proteomic and Metabolomic Characterization of COVID-19 Patient Sera. Cell 2020, 182, 59–72.e15.
- Yang, J.; Chen, C.; Chen, W.; Huang, L.; Fu, Z.; Ye, K.; Lv, L.; Nong, Z.; Zhou, X.; Lu, W.; et al. Proteomics and metabonomics analyses of COVID-19 complications in patients with pulmonary fibrosis. Sci. Rep. 2021, 11, 14601.
- Caterino, M.; Ruoppolo, M.; Villani, G.R.D.; Marchese, E.; Costanzo, M.; Sotgiu, G.; Dore, S.; Franconi, F.; Campesi, I. Influence of Sex on Urinary Organic Acids: A Cross-Sectional Study in Children. Int. J. Mol. Sci. 2020, 21, 582.
- Melo, M.G.; Remacle, N.; Cudré-Cung, H.-P.; Roux, C.; Poms, M.; Cudalbu, C.; Barroso, M.; Gersting, S.W.; Feichtinger, R.G.; Mayr, J.A.; et al. The first knock-in rat model for glutaric aciduria type I allows further insights into pathophysiology in brain and periphery. Mol. Genet. Metab. 2021, 133, 157–181.
- Costanzo, M.; Fiocchetti, M.; Ascenzi, P.; Marino, M.; Caterino, M.; Ruoppolo, M. Proteomic and Bioinformatic Investigation of Altered Pathways in Neuroglobin-Deficient Breast Cancer Cells. Molecules 2021, 26, 2397.
- Costanzo, M.; Caterino, M.; Salvatori, I.; Manganelli, V.; Ferri, A.; Misasi, R.; Ruoppolo, M. Proteome data of neuroblastoma cells overexpressing Neuroglobin. Data Brief 2022, 41, 107843.
