Papillomaviruses are a large family of non-enveloped viruses with ~8000 base pair, circular, double stranded DNA genomes. They have been detected in almost all vertebrates, are highly host-specific and preferentially infect squamous epithelial tissues. Infections with high-risk human papillomaviruses cause ~5% of all human cancers. E6 and E7 are the only viral genes that are consistently expressed in cancers, and they are necessary for tumor initiation, progression, and maintenance. E6 and E7 encode small proteins that lack intrinsic enzymatic activities and they function by binding to cellular regulatory molecules, thereby subverting normal cellular homeostasis. Much effort has focused on identifying protein targets of the E6 and E7 proteins, but it has been estimated that ~98% of the human transcriptome does not encode proteins. Long noncoding RNAs (lncRNAs) are defined as transcripts of >200 nucleotides with no or limited coding potential of <100 amino acids. There is a growing interest in studying noncoding RNAs as biochemical targets and biological mediators of human papillomavirus (HPV) E6/E7 oncogenic activities.
- human papillomavirus
- viral oncogenesis
- cervical carcinoma
- lncRNA
- E6
- E7
1. Human Papillomaviruses as Oncogenic Drivers
2. Long Noncoding RNAs

3. Deregulation of lncRNAs in Cervical Carcinomas
|
lncRNA |
|---|
|
lncRNA |
Oncogenic Phenotype |
Oncogenic Phenotype Proposed Mechanism |
References |
||||
|---|---|---|---|---|---|---|---|
Proposed Mechanism |
Reference |
||||||
|
ANRIL |
|||||||
|
GAS5 |
Proliferation, migration, invasion |
Proliferation, invasion, migration PI3K/AKT; Cyclin D1, CDK4, CDK6, N-cadherin, Vimentin expression |
E-cadherin, Vimentin |
||||
[ | ][76] |
ARAP1-AS1 |
Proliferation, invasion |
MYC translation by PSF/PTB |
|||
|
Proliferation, migration, invasion, colony formation |
miR-21 expression/STAT3 | ||||||
[ | ][77] |
BLACAT1 |
Proliferation, migration, invasion |
||||
|
Radiosensitivity |
WNT signaling/β-catenin |
||||||
miR-106b sponging/IER3 |
CCAT2 |
Proliferation, apoptosis |
None reported |
[ | |||
|
HOTAIR |
Decreased polycomb repression |
Binding to HPV E7 | |||||
[ | ][79] |
CCEPR (CCHE1) |
|||||
|
Lnc-CCDST |
Proliferation |
PCNA mRNA stabilization |
[31 |
Migration, invasion, angiogenesis ][32] |
|||
DHX9, MDM2 scaffolding | |||||||
|
MEG3 |
Proliferation |
Proliferation, apoptosis independent of PCNA mRNA |
Binding, degradation of P-STAT3 |
||||
[ | ][81] |
CRNDE |
Proliferation, migration, invasion |
||||
|
Proliferation, colony formation, apoptosis | miR-183 sponging/cyclin B1 |
miR-21-5p expression/TP53 |
|||||
[ | ][82] |
Proliferation |
|||||
|
STXBP5-AS1 | PUMA expression |
Viability, invasion |
|||||
miR-96-5p expression/PTEN |
DANCR |
Proliferation, migration, invasion |
|||||
|
TINCR | miR-665 sponging/TGFβ-R1-ERK-SMAD |
Differentiation, colony formation, migration [35][ |
S100A8 and other ZNF750 targets 36] |
||||
[ | ][84] |
||||||
|
WT1-AS |
Proliferation, migration, invasion, epithelial to mesenchymal transition (EMT) |
Proliferation miR-335-5p sponging/ROCK1 |
TP53 |
||||
[ |
EBIC (TMPOP2) |
Motility, invasion |
E-cadherin silencing by EZH2 |
[37 | |||
|
Proliferation, invasion, migration | ] | [ |
miR-203a-5p binding/FOXN2 38] |
||||
[ | ][86] |
Proliferation |
miR-375, miR-139 sponging HPV E6/E7 expression |
||||
|
XLOC_010588 |
Proliferation |
MYC mRNA binding/degradation |
FAM83H-AS1 |
Proliferation, migration and apoptosis |
G1/S-phase transition |
||
|
GATA6-AS |
Migration, invasion |
MTK-1 |
|||||
|
H19 |
Proliferation, anchorage independent growth |
None reported |
|||||
|
HOTAIR |
Apoptosis, invasion, migration |
NOTCH signaling |
|||||
|
Apoptosis, proliferation, invasion |
miR-23b sponging/MAPK1 axis |
||||||
|
Autophagy, EMT |
WNT signaling |
||||||
|
Proliferation |
miR-143-3p sponging/BCL2 |
||||||
|
HOXD-AS1 |
Proliferation |
Ras/ERK |
|||||
|
Linc00483 |
Proliferation, apoptosis, invasion, migration |
miR-508-3p sponging/RGS17 |
|||||
|
LINP1 |
DNA damage repair (Non-homologous end joining) |
KU80, DNA-PKcs binding |
|||||
|
Lnc-IL7R |
Apoptosis |
BCL2/caspase 3 |
|||||
|
LUCAT1 |
Proliferation, migration, invasion |
miR-181a sponging |
|||||
|
MALAT1 |
Cell invasion and metastasis |
inhibition of EMT genes |
|||||
|
Proliferation, migration, invasion |
miR-625-5p/AKT2 |
||||||
|
Proliferation |
Mir-625-5p/NF-kB signaling |
||||||
|
Cisplatin resistance |
PI3K/AKT |
||||||
|
MIR205HG |
Proliferation, apoptosis, migration |
SRSF1/KRT17 axis |
|||||
|
NEAT1 |
Proliferation, invasion |
PI3K/AKT |
|||||
|
Colony formation, migration, invasion |
miR-133a sponging/SOX4 |
||||||
|
NORAD |
Proliferation, invasion |
miR-590-3p sponging/SIP1 |
|||||
|
PANDAR |
Proliferation |
None reported |
|||||
|
PVT1 |
Proliferation, invasion |
Inhibiting TGFβ; miR-140-5p sponging/SMAD3 |
|||||
|
EMT, chemoresistance |
miR-195 epigenetic silencing |
||||||
|
SNHG8 |
Proliferation, apoptosis |
RECK silencing by EZH2 |
|||||
|
SNHG12 |
Proliferation, apoptosis |
ERK/Slug |
|||||
|
SNHG16 |
Proliferation, invasion |
PARP9 expression by SPI1 binding |
|||||
|
TUG1 |
Proliferation, apoptosis, invasion, tumor growth |
miR-138-5p sponging/SIRT1 |
|||||
|
Proliferation, apoptosis, EMT |
BCL-2, caspase 3; fibronectin, vimentin, and cytokeratin |
||||||
|
TP73-AS1 |
Proliferation, migration |
miR-329-3p sponging/SMAD2 |
|||||
|
Proliferation, migration, invasion |
miR-607 sponging/CCND2 |
||||||
|
UCA1 |
Radioresistance |
HK2/glycolytic pathway |
|||||
|
XIST |
Proliferation |
miR-140-5p sponging/ORC1 |
|||||
|
Proliferation, invasion, apoptosis, EMT |
miR-200a sponging/FUS |
||||||
|
ZEB-AS1 |
Proliferation, migration, invasion, EMT |
ZEB1 expression |
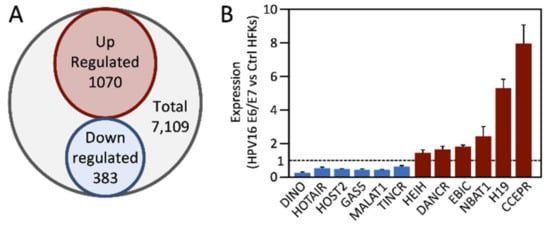
4. Deregulation of lncRNAs by HPV E6 and/or E7 Proteins