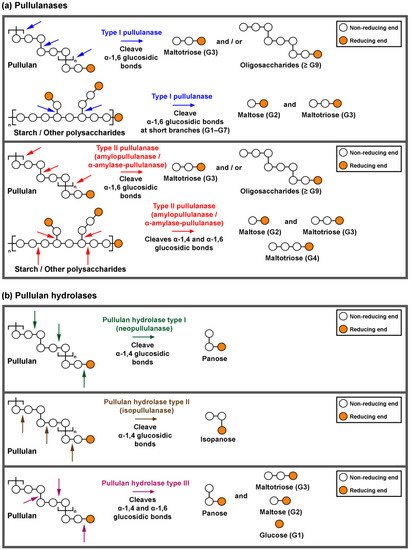
Starch and pullulan degrading enzymes are essential industrial biocatalysts. Pullulan-degrading enzymes are grouped into pullulanases (types I and type II) and pullulan hydrolase (types I, II and III).
- amylopullulanase
- alpha-amylase-pullulanase
- carbohydrate-active enzyme
1. Classification and Action of Pullulan-Degrading Enzymes
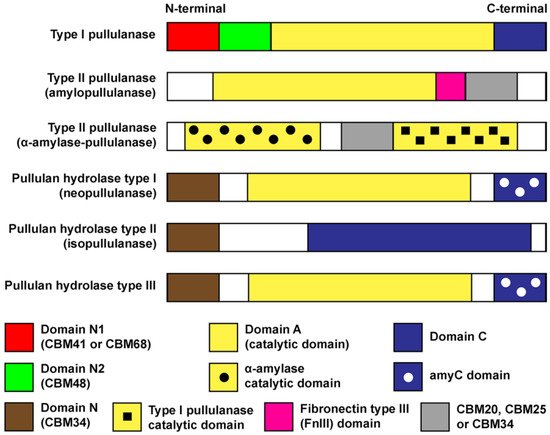
2. Domains, Structures and Properties of Pullulan-Degrading Enzymes
3. Single Immobilisation of Pullulan-Degrading Enzyme
4. Co-Immobilisation of Pullulan-Degrading Enzymes
- (i) free enzymes mixture (PulASK+TASKA)(i)
-
free enzymes mixture (PulASK+TASKA)
- (ii)
-
individual immobilised enzymes mixture (HFA403/M–PulASK+ HFA403/M–TASKA)
- (ii) individual immobilised enzymes mixture (HFA403/M–PulASK+ HFA403/M–TASKA)(iii)
- (iii) co-immobilised enzymes (PulASK–HFA403/M–TASKA).
-
co-immobilised enzymes (PulASK–HFA403/M–TASKA).The accumulated data from that setup indicated that the total amount and the generated spectrum of reducing sugars using (iii) co-immobilised enzymes were significantly different from either the (i) free enzymes mixture or (ii) the individual immobilised enzymes mixture [86]. rWesearchers thought that structural changes in PulASK and TASKA may alter the substrate-enzyme-product dynamic interaction and thus drastically modify their product specificities. Nevertheless, due to random covalent attachment, that work could not pinpoint the exact residues bound to the carrier, or how these residues affect the product specificity of the bound enzyme.
5. Protein Engineering for Improving the Performance of Pullulan-Degrading Enzymes
Protein engineering, also known as protein mutagenesis, is an approach that alters or improves enzyme biochemical features (i.e., substrate specificity, product specificity, specific activity, reducing inhibition effects or thermostability). The strategies include (i) directed evolution (error-prone PCR, staggered extension PCR, DNA shuffling, etc.) and (ii) rational design or site-directed mutagenesis (mainly mega primer PCR and overlap extension PCR methods) [117]. As expected, there are relatively fewer attempts for pullulan hydrolases engineering [37,118,119]. One central mutagenesis theme for pullulan-degrading enzymes is understanding the role of a residue at a particular position, specific amino acids stretch, motif or domain. For instance, Kim et al. [61] and Li et al. [126] performed CBM truncation to understand starch saccharification function. In another work, Kahar et al. elucidated that a short loop of the native protein prevents the native enzyme from generating reducing sugars from short linear or branched oligosaccharides [135]. In one particular study, Chen et al. shifted optimum pH from 5.0 to 4.0, and the mutant pullulanase exhibited an increased tolerance against acid denaturation [136]. Using pullulanase as a role model, the concept of ECSM (evolutionary coupling saturation mutagenesis) seems promising in cutting the time in identifying the amino acid positions for mutations [137]. The ECSM is an extension concept of an open-source software EVcouplings that perform computational predictions based on evolutionary sequence covariation (https://evcouplings.org). Improving activity half-life or structural stability is another central theme for pullulanase. For instance, among the six constructed variant proteins, pullulanase mutant G692M from Geobacillus thermoleovorans had a two-fold improvement in the activity half-life [127]. In the past decade, several computation predictors were created to assist researchers in doing in silico protein structure stabilisation analysis before starting to construct variants on the lab bench. Most of these predictors calculate ΔΔG (changes in the Gibbs free energy) between in silico native and variant proteins. In early times, these predictors applied the thermodynamic information recorded in the ProTherm database [140]. Approximately 20 years ago, ProTherm contained less than 6000 data entries and in 2013, the database updated to >25,000 thermodynamic data. Structure-based predictors, including FoldX, I-Mutant series, CUPSAT, PoPMuSiC, STRUM and etc., may outperform sequence-based predictors [141]. For the mutant pullulanase G692M that rwesearchers stated at the beginning of this paragraph, the researchers utilised FoldX, I-Mutant 3.0 and dDFIRE [127]. From structural biologists’ perspective, X-ray protein structure is more accurate than homology-modelling predicted models. One may face drawbacks if the input files for structure-based predictors are the protein models instead of high-resolution X-ray structures. Crystallography is costly and the state-of-the-art AlphaFold may be an option to generate a better model structure.More recent predictors apply protein structural datasets and evolutional information to train machine learning. A few years ago, a research team noticed several problems with ProTherm, and they later cleaned the database before developing the PON-tstab predictor [148]. To overcome other limitations of ProTherm, the developers for DDGun and DDGun3D used a method that combines anti-symmetric features for predicting the ΔΔG upon variation, and claimed that this untrained and straightforward method has excellent performance. So far, only a few predictors address the anti-symmetric issue, and examples of these programs include PROTS-RF, INPS, SDM, ProTstab and a few others [149]. Predictor DeepDDG, a neural network approach, was trained using ProTherm, and manually curated literature data. The developer suggested that DeepDDG outperformed the other eleven commonly used predictors [146]. The identical research group that created PON-tstab later proposed a new platform, ProTstab, which used a >3500 proteolysis and mass spectrometry (LiP-MS) dataset to train the gradient-boosting based machine learning platform. Different algorithms may suggest contradictory suggestions, and it is time-consuming to try one predictor at a time. One may consider iStable 2.0, an integrated platform consisting of eleven structure- and sequence-based tools [147]. Readers can refer to theis excellent review that compares the limitations and challenges of each predictor from the angle of future applications in precision medicine [149]. rWesearchers observed that not many pullulanase protein engineers use the tools mentioned above; therefore, keywords stated in this subsection will not appear in bibliometric records (Figure 1c).
6. Influence of Next-Generation Metagenome Sequencing on Novel Pullulan-Degrading Enzymes Discovery
Next-generation sequencing (NGS) is a powerful tool in natural science and biotechnology in the current era. Metagenome sequencing with computation pipelines can generate a tremendous amount of novel macromolecule sequences. Interested readers in this topic can refer to these publications for more insights [150,151,152,153,154]. resWearchers searched using the keyword ‘metagenome and X’ (where X is cellulase, lipase, amylase or pullulanase), the hit of research articles in the Scopus database is 180, 137, 60, 5, respectively. The articles related to pullulanase are listed in the references [155,156,157,158,159]. No relevant documents for pullulan hydrolase types I, II and III were found.A type I pullulanase derived from Reshi hot spring metagenome dataset was reported in 2021 [157]. The enzyme, named PulM, has very low sequence similarity to currently characterised pullulanases. For instance, PulM shares only 42.9% identity to a type I pullulanase from Geobacillus stearothermophilus. PulM had an optimum catalytic temperature at 40 °C. Interestingly, the enzyme remained 50% active at 4 °C, a relatively higher activity than other pullulanases obtained from isolated bacteria sampled from hot springs [157]. In another recent article, a novel type I pullulanase (PersiPul1) was established from cow rumen metagenomic data [156]. The enzyme is active from 30–80 °C and retained close to 60% of its original activity at 80 °C. In the same study, the researchers blended a cocktail of type I pullulanase (PersiPul1) and α-amylase PersiAmy2 (also derived from metagenome strategy, [160]). They used the enzyme cocktail to improve bread physical and sensory properties [156].Amylopullulanase PulSS4 was a type II pullulanase derived via the metagenome approach [159]. The gene encoding this enzyme was identified in a gut metagenome (clonal functional library approach) of Hermetia illucens (black soldier fly). PulSS4 closest counterpart (51%) is a protein from the anaerobic bacterium Amphibacillus xylanus. The authors claimed that PulSS4 had the maximum activity at pH 9.0 and exhibited a broad pH tolerance [159].The laboratory recently performed Illumina shotgun sequencing using environmental DNA extracted from biofilms attaching to plant litters socked in a local hot spring. Using the assembled contigs (Figure 3c), researchers have mined >10,000 sequences related to various CAZymes and identified close to 40 sequences for pullulan-degrading enzymes. All of them are type I or type II pullulanases, except one was detected as neopullulanase. This may suggest that pullulan hydrolases are rarely encoded in the genomes of thermophiles. Nevertheless, this approach enables reusearchers to find novel pullulanase sequences from rare prokaryotes, for instance, Candidatus Sericytochromatia and Candidatus Roseilinea. Ouresearcher's team is currently analysing the high-quality genomes generated by metagenome-assembled genomes (MAGs) using short-reads from Illumina. Besides, Oxford Nanopore and HiFi PacBio long-reads sequencing will soon become a common practice for MAGS. The MAGs strategy might extend researcher'sour ability to mine novel pullulan-degrading enzymes.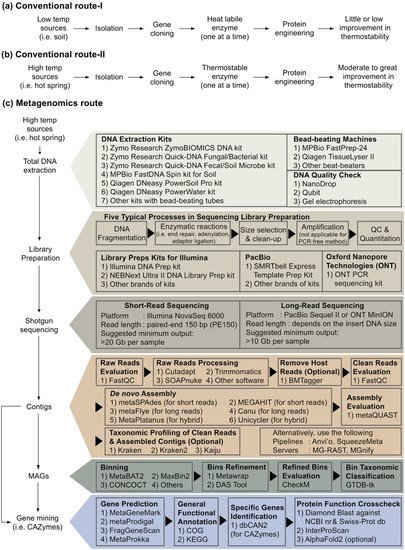 Figure 3. Research flow chart to find novel active and thermostable pullulan-degrading enzymes.Research flow chart to find novel active and thermostable pullulan-degrading enzymes.---------------------------------------------------------------------Suppose searching for active and thermostable pullulan-degrading enzymes is the aim of ouresearcher's research. Despite the feasibility of protein engineering approach to design enzymes with improved thermostability and longer half-life, especially among the thermolabile origins, the engineered enzymes might be far from the temperature threshold required by the starch- and pullulan processing industries (Figure 3a). Harness native biocatalyst from a large pool of metagenome-derived enzymes, particularly from samples such as high-temperature hot springs, is perhaps a better route than the protein engineering approach (Figure 3c). Biologists may be unacquainted with the advanced bioinformatic tools summarised in Figure 3c. In addition, from ouresearcher's own experience and the limited examples of pullulan-degrading enzymes [155,156,157,158,159], metagenome-derived enzymes may exhibit low sequence similarity to patented sequences.
Figure 3. Research flow chart to find novel active and thermostable pullulan-degrading enzymes.Research flow chart to find novel active and thermostable pullulan-degrading enzymes.---------------------------------------------------------------------Suppose searching for active and thermostable pullulan-degrading enzymes is the aim of ouresearcher's research. Despite the feasibility of protein engineering approach to design enzymes with improved thermostability and longer half-life, especially among the thermolabile origins, the engineered enzymes might be far from the temperature threshold required by the starch- and pullulan processing industries (Figure 3a). Harness native biocatalyst from a large pool of metagenome-derived enzymes, particularly from samples such as high-temperature hot springs, is perhaps a better route than the protein engineering approach (Figure 3c). Biologists may be unacquainted with the advanced bioinformatic tools summarised in Figure 3c. In addition, from ouresearcher's own experience and the limited examples of pullulan-degrading enzymes [155,156,157,158,159], metagenome-derived enzymes may exhibit low sequence similarity to patented sequences.
