Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Pinar Uysal-Onganer and Version 5 by Bruce Ren.
Pancreatic ductal adenocarcinoma (PDAC) is the most common and aggressive type of pancreatic cancer (PCa) with a low survival rate. microRNAs (miRs) are endogenous, non-coding RNAs that moderate numerous biological processes. miRs have been associated with the chemoresistance and metastasis of PDAC and the presence of a subpopulation of highly plastic “stem”-like cells within the tumor, known as cancer stem cells (CSCs).
- pancreatic ductal adenocarcinoma
- microRNAs
- non-coding RNAs
- cancer stem cells
- metastasis
- epithelial–mesenchymal transition
1. Introduction
Pancreatic ductal adenocarcinoma (PDAC) is the eighth primary source of cancer-related deaths globally with a 5-year survival rate of 3–6% [1][2][3][4][1,2,3,4]. Approximately, 10–20% of PDAC patients are compatible for surgery at the time of diagnosis, and 9.7% of PDAC cases are at a local stage when initially diagnosed [5]. microRNAs (miRs) are 18 to 24 nucleotides-long, endogenous, non-coding, evolutionarily conserved, single-stranded RNA molecules. miRs can moderate gene expression at the posttranscriptional level through the binding to the complementary sequences of their target mRNAs at the 3′ untranslated regions (UTRs), which allow them to control the expression levels of several genes and regulate various signaling pathways [6][7][8][9][6,7,8,9]. Preliminary studies have suggested a correlation between aberrant expression levels of numerous miRs with PDAC [10][11][12][13][10,11,12,13]. miRs can act as oncogenic miRs (oncomiRs) or tumor suppressor miRs. Especially in PDAC, miR-21, miR-155, and miR-221 have been found to act as oncomiRs, while miR-126 and miR-375 were shown to act as tumor suppressors miRs [14][15][14,15]. miR-21, miR-221, and miR-155 can distinguish cases of PDAC from healthy individuals with a sensitivity of approximately 64% and a specificity of 89% [16][17][16,17]. Importantly, miRs present a higher sensitivity as a diagnostic marker than the current diagnostic marker carbohydrate antigen (CA 19-9), especially for the early diagnosis of PDAC [18][19][20][21][18,19,20,21].
Cancer stem cells (CSCs) are involved in chemoresistance and play critical roles in the metastasis of several cancers, including PDAC [22][23][24][25][26][27][28][29][22,23,24,25,26,27,28,29]. CSCs contribute to elevated expression levels of anti-apoptotic proteins, ABC transporters, and multidrug resistance genes, high autophagic flux that leads to microenvironment stresses [30][31][32][33][34][30,31,32,33,34]. Pancreatic CSCs (PCSCs) are less than 1% of all pancreatic cancer cells and are critical mediators of PDAC tumor growth, maintenance, metastasis, and chemoresistance [35]. EMT is characterized as a critical mechanism of the metastatic cascade, which includes the loss of cell adhesion, elevated cell motility, the repression of E-cadherin, and the upregulation of mesenchymal markers, such as Vimentin, N-cadherin, Snail, and Zeb1 [36]. E-cadherin downregulation is associated with poor prognosis, differentiation, and chemoresistance in PDAC [37][38][39][40][37,38,39,40]. Transcription marker Zeb1 suppresses E-cadherin through the repression of both miR-203 (an inhibitor of stemness) and miR-200 family members, which control the expression levels of stem cell factors [41]. Zeb1 overexpression is linked to advanced PDAC stages and poor malignancy outcome, migration, and invasion in response to nuclear factor kappa-light-chain-enhancer of activated B cells (NF-κB) signaling [42][43][44][42,43,44]. Non-canonical Wnt-11 overexpression is associated with poor prognosis and tumor-node-metastasis (TNM) staging in PDAC [45][46][47][48][49][45,46,47,48,49].
Higher expression levels of Snail, a potent EMT-inducing transcription factor, are related to 80% of PDAC cases, E-cadherin downregulation, lymph node invasion, higher tumor grade, and poorly differentiated PDAC cells [50]. EMT is regulated via molecular pathways linked to oncogenic and tumor suppressor non-coding RNAs, chromatin remodeling, epigenetic and posttranslational modifications, alternative splicing events, and protein stability [51][52][51,52]. Vimentin is an essential marker of EMT and associated with Notch and miR-200 expression levels and it can affect treatment response in vitro, including elevated gemcitabine-resistance in PDAC [53][54][53,54]. Overexpression of Vimentin is linked to metastasis and poor overall survival in PDAC [55][56][55,56]. Furthermore, the association between EMT and CSCs has been extensively evaluated, for instance, PDAC cells, which have undergone EMT, and express epithelial markers, such as E-cadherin and mesenchymal markers, such as Zinc Finger E-Box Binding Homeobox 1 (Zeb1), and Snail exhibit stem cell properties [57][58][57,58]. The main markers of PCSCs are CD133, CD24, CD44, ESA/EpCAM (epithelial-specific antigen), c-Met, ALDH1, DclK1, CXCR4, and Lgr5 [30][59][60][30,59,60]. Importantly, EMT and autophagy processes are also closely linked with CSCs markers during PDAC development [59][60][59,60]. Recent reports stated that CD24, CD44, CXCR4, ESA, and nestin are upregulated in advanced pancreatic intraepithelial neoplasia (PanIN) grades [61], while others have shown that the expression of cMet+ CD133+ CD34+ CD45− Ter119−, Pdx1, CD9, CD24, CD44, CD13, and CD133, are linked to poor prognosis of PDAC [61][62][63][64][65][66][67][61,62,63,64,65,66,67].
2. MicroRNA-21 Regulates Stemness in Pancreatic Ductal Adenocarcinoma Cells
In summary, using three different PDAC cell lines, thwe researchers found that miR-21, miR-221, miR-155, and miR-126 expressions were significantly altered in MiaPaca-2, Panc-1, and BxPC3 PDAC cell lines, compared with normal pancreatic ductal epithelial cell lines (HPDE). Following the knockout of miR-21 in Panc-1 and MiaPaca-2 cells using CRISPR-Cas9, reversed expressions of E-cadherin, Vimentin, Snail, Wnt-11, and Zeb1 were detected, suggesting that these markers are targets of miR-21. Expression levels of the CSC markers, such as CD133, CD44, CD24, CXCR4, and ALDH1, were significantly downregulated depending on miR-21 status. KO of miR-21 led to a significant reduction in cellular invasiveness of Panc-1 and MiaPaCa-2 cells and a significant decrease in cellular proliferation of MiaPaCa-2 cells. Overall, the researchersour data suggest that miR-21 is involved in the pathophysiology of PDAC.
2.1. Expression Profiles of miR-21, miR-221, miR-155, and miR-126 in PDAC In Vitro
PDAC associated miR-21, miR-155, miR-221, miR-155, and miR-126 expression levels were quantified using RT-qPCR in three different PDAC cell lines (MiaPaCa-2, Panc-1, and BxPC3), compared with human pancreatic ductal epithelial (HPDE), which served as the control cell line. The HPDE cells had the most epithelial properties expressed relatively, with the least miR-21-5p, miR-155-5p, and miR-221-5p and this was used as a ‘baseline’ (Figure 1A–C). Out of the cell lines tested, Panc-1 cells expressed the highest levels of miR-21-5p (117-fold elevation; n = 3; p < 0.0001; Figure 1A), miR-155-5p (105-fold increase; n = 3; p < 0.0001; Figure 1B), and miR-221-5p (32-fold upregulation; n = 3; p < 0.0001; Figure 1C) relatively compared with HPDE cells. Similarly, MiaPaCa-2 cells presented a 43-fold increase in the miR-21-5p (n = 3; p < 0.05; Figure 1A), a 51-fold overexpression in miR-155-5p (n = 3; p < 0.01; Figure 1B) and a 21-fold significant rise in miR-221-5p (n = 3; p < 0.001; Figure 1C). BxPC3 cells did not show significant changes in both miR-21-5p and miR-221-5p expressions (16-fold; 3-fold, respectively; n = 3; p > 0.05 for both; Figure 1A,C), however, they showed a 36-fold significant upregulation in miR-155-5p expression (n = 3; p < 0.05; Figure 1B). The expression levels of miR-126-5p indicated a 57-fold downregulation in BxPC3 cells (n = 3; p < 0.01; Figure 1D), a 81-fold decrease in the MiaPaCa-2 cell line (n = 3; p < 0.001; Figure 1D), and a 94-fold reduction in Panc-1 cells (n = 3; p < 0.0001; Figure 1D) in comparison with normal HPDE cells.
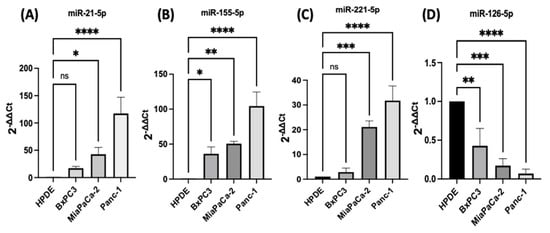

Figure 1. RT-qPCR analysis of miRs levels miR-21, miR-221, miR-155, and miR-126 in PDAC cell lines (BxPC3, MiaPaCa-2, Panc-1, compared with HPDE). (A) miR-21-5p relative expression was significantly increased in MiaPaCa-2 and Panc-1 but not in BxPC3 PDAC cell lines. (B) miR-155-5p relative expression was significantly overexpressed in BxPC3, MiaPaCa-2, and Panc-1 PDAC cell lines. (C) miR-221-5p relative expression was significantly upregulated in MiaPaCa-2 and Panc-1 but not in BxPC3. The column graphic represents the average of three replicates of RNA isolated from each cell line. (D) miR-126-5p relative expression was significantly reduced in PDAC cell lines. The column graphs represent the average of three replicates of RNA isolated from each cell line. Data normalized according to RNU6 expression by fold analysis (n = 3, p < 0.05 for all). p-values are indicated as: * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001; error bars indicate standard deviation (SD).
2.2. Development of miR-21 KO MiaPaca-2 and Panc-1 Cell Lines
The Ouresearchers qRT-PCR data showed that miR-21 expression was the most elevated in the Panc-1 and MiaPaCa-2 PDAC cell lines correlating with in vivo data. Therefore, both Panc-1 and MiaPaca-2 were used to further investigate the effect of knocking out miR-21, and cells were transduced with four different miR-21 gRNAs, as well as a control vector. miR-21 expression analysis showed that miR-21 expression was significantly reduced by 65 and 73-fold in Panc-1 knockout (KO) clones 2 (KO2) and 4 (KO4), respectively, compared with Panc-1 vector alone (n = 3; p < 0.0001 for all; Figure 2A). Similarly, expression levels of miR-21 were also significantly decreased by 83-fold in KO2 and 97-fold in KO4 (n = 3; p < 0.01 for all; Figure 2B) in MiaPaCa-2. The knockout miR-21 Panc-1 and MiaPaCa-2 PDAC cell lines were further assessed for EMT, and CSC markers and cellular invasion.
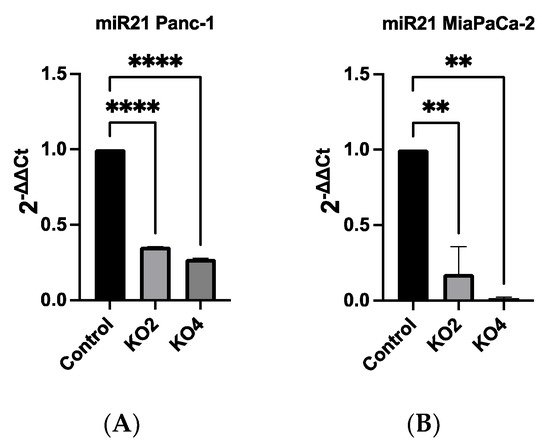

Figure 2. miR-21 expression levels are significantly reduced in Panc-1 and MiaPaCa-2 Kos. (A) Relative expression levels of miR-21 in miR-21 KO Panc-1 cells. miR-21 expression levels of miR-21 knockout clone 2 (KO2) and miR-21 knockout clone 4 (KO4) cell colonies were downregulated significantly compared to untreated control Panc-1 cells (n = 3; p < 0.0001 for all). The column graphic represents the average of three replicates of RNA isolated from the Panc-1 control and its Kos. Data normalized according to RNU6 expression by fold analysis (n = 3, p < 0.05). (B) Relative expression levels of miR-21 in miR-21 KO MiaPaCa-2 cells. miR-21 expression levels of miR-21 knockout clone 2 (KO2) and miR-21 knockout clone 4 (KO4) cell colonies were reduced significantly compared to untreated control MiaPaCa-2 cells (n = 3; p < 0.01 for all). The column graphic represents the average of three replicates of RNA isolated from MiaPaCa-2 control and its Kos. Data normalized according to RNU6 expression by fold analysis (n = 3, p < 0.05). p-values are indicated as: ** p ≤ 0.01; **** p ≤ 0.0001; error bars indicate SD.
2.3. miR-21 Kos Reduce Expression Levels of EMT-Related Markers
The effects of miR-21 KO on regulation of key EMT was assessed using miR-21 KO in Panc-1 and MiaPaCa-2 PDAC cells. E-cadherin mRNA expression levels were found to be significantly overexpressed by 932-fold in Panc-1 miR-21 KO2 and by 906-fold in KO4 (n = 3; p < 0.05 and 0.01, respectively; Figure 3A). Vimentin mRNA levels decreased by 96-fold in KO2 and 99-fold in KO4 (n = 3; p < 0.0001 for all; Figure 3A), whereas Snail mRNA levels were 100-fold reduced in both KO2 and KO4 (n = 3; p < 0.0001 for all; Figure 3A). Zeb1 mRNA expression was reduced by 40-fold in KO2 and by 46-fold in KO4 (n = 3; p < 0.01 for all; Figure 3A) in comparison to the Panc-1 vector alone. The Wnt-11 mRNA expression level was significantly reduced in Panc-1 miR-21 KO4 (58-fold; n = 3; p < 0.05; Figure 3A), but there was not a significant reduction in KO2 (35-fold, n = 3; p > 0.05; Figure 3A). Similarly, in the MiaPaca-2, E-cadherin mRNA levels were significantly upregulated by 349-fold in KO2 (n = 3; p < 0.01; Figure 3B) and by 476-fold in KO4 (n = 3; p < 0.001; Figure 3B). Additionally, vimentin mRNA levels were decreased by 71-fold in KO2 and by 99-fold in KO4 (n = 3; p < 0.01 and 0.001, respectively; Figure 3B), while Snail mRNA levels presented a significant decrease in KO2 (69-fold) and KO4 (80-fold) (n = 3; p < 0.05 for all; Figure 3B). Zeb1 mRNA levels were suppressed significantly by 49 and 77-fold in KO2 and KO4, respectively (n = 3; p < 0.01 and 0.001, respectively; Figure 3B), and Wnt-11 mRNA expression levels reduced in both KO2 and KO4 (57-fold and 49-fold, respectively; n = 3; p < 0.01 for all; Figure 3B). According to immunofluorescence results, E-cadherin was upregulated; Snail and Wnt-11 were downregulated in miR-21 KO clones when compared to control (n = 3; Figure 3C). Overall, the ouresearchers data suggest that miR-21 is involved in EMT through suppressing epithelial characteristics in the cells.
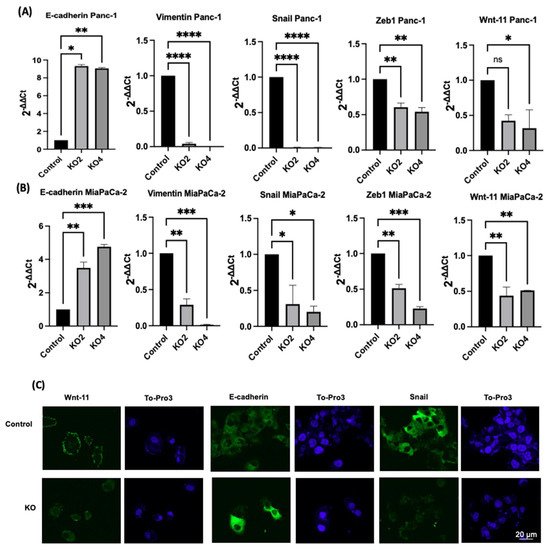

Figure 3. The silencing of miR-21 reduced the mesenchymal phenotype of PDAC cells. (A) E-cadherin, Vimentin, Snail, Wnt-11, and Zeb1 in the Panc-1 cell line relative mRNA expression levels. Relative mRNA expression levels of Vimentin, Snail, and Zeb1 were significantly downregulated in miR-21 KO2 and KO4 in the Panc-1 cell line, while E-cadherin was significantly upregulated. Relative mRNA expression levels Wnt-11 were significantly reduced in miR-21 KO4 but not in miR-21 KO2 in the Panc-1 cell line. The column graphic represents the average of three replicates of RNA isolated from Panc-1 control and its Kos. Data normalized according to RNA polymerase II (RPII) expression by fold analysis (n = 3, p < 0.05). (B) Relative mRNA expression levels of E-cadherin, vimentin, Snail, Wnt-11, and Zeb1 in the MiaPaCa-2 cell line. Relative mRNA expression levels of vimentin, Snail, Zeb1, and Wnt-11 significantly decreased in miR-21 KO2 and KO4 in the MiaPaCa-2 cell line, while E-cadherin significantly increased. The column graphic represents the average of three replicates of RNA isolated from MiaPaCa-2 control and its Kos. Data normalized according to RNA polymerase II (RPII) expression by fold analysis (n = 3, p < 0.05). Exact p-values are indicated (* p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001); error bars indicate SD. (C) Immunofluorescence assays were performed to show Wnt-11, E-cadherin, Snail protein levels, and protein localization (green) in Panc-1 wt (Control) and miR-21 KO cells. To-Pro3 (blue) was used for staining nuclei. KO2 immunofluorescence results are shown as a representative to compare to control (n = 3), the scale bar represents 20 μm in all images.
2.4. miR-21 Kos Diminish Expressions of CSC Markers in PDAC
The expression levels of specific selected CSCs, which are involved in PDAC progression, were assessed in the miR-21 PDAC KO cell lines generated from both Panc-2 and MiaPaCa-2; this included CSC markers CD44, CD24, CD133, CXCR4, and ALDH1. In Panc-1 miR-21 KO clones 2 and 4, the mRNA expression levels of CD44 were decreased by 97% (n = 3; p < 0.01 for all; Figure 4A), whereas CD133 was downregulated by 76% in KO2 and by 52% in KO4 (n = 3; p < 0.01 and p < 0.05, respectively; Figure 4A). CXCR4 was reduced by 86% in KO2 and by 100% in KO4 (n = 3; p < 0.0001 for all; Figure 4A), whereas ALDH1 expression levels were reduced by 77% in KO2 and by 95% in KO4 (n = 3; p < 0.01 for all; Figure 4A). The mRNA expression levels of CD24 were reduced by 77% in KO2 and by 88% in KO4 (n = 3; p < 0.05 and p < 0.05, respectively; Figure 4A), compared with the control Panc-1 vector alone. In the MiaPaCa-2 miR-21 knockout cells, CD44 mRNA expression levels decreased significantly by 75% in KO2 (n = 3; p < 0.001; Figure 4B) and by 40% in KO4 (n = 3; p < 0.01; Figure 4B), whereas CD133 mRNA levels were decreased by 50% in KO2 and by 42% in KO4 (n = 3; p < 0.05 for all; Figure 4B). Similarly, CXCR4 mRNA expression levels were reduced by 51% in KO2 and by 69% in KO4 (n = 3; p < 0.01 for all; Figure 4B), while ALDH1 mRNA levels decreased by 66% in KO2 and 46% in KO4 (n = 3; p < 0.001 for all; Figure 4B). The mRNA expression levels of CD24 were significantly reduced by 73% in KO2 (n = 3; p < 0.01; Figure 4B) and by 53% in KO4 (n = 3; p < 0.05; Figure 4B). Hence, the findings show that through the knocking out of miR-21 in two PDAC cell lines, the stemness markers CD44, CD133, CD24, CXCR4, and ALHD1 were significantly reduced in both cell lines assessed.
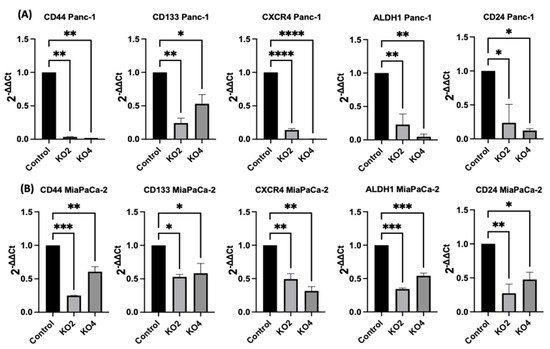

Figure 4. Expression of CSCs markers in PDAC cell lines following miR-21 knockout. (A) mRNA expression levels of CD44, CD133, CXCR4, ALDH1, and CD24 in the Panc-1 miR-21 KO cell line, compared with the Panc-1 vector alone. Relative mRNA expression levels of CD44, CD133, CXCR4, ALDH1, and CD24 are significantly reduced in miR-21 KO Panc-1 cells. The column graphic represents the average of three replicates of RNA isolated from Panc-1 control and its Kos. Data normalized according to RPII expression by fold analysis (n = 3, p < 0.05). (B) mRNA expression levels of CD44, CD133, CXCR4, ALDH1, and CD24 in the miR-21 KO MiaPaCa-2 cell line, compared with MiaPaCa-2 vector alone. Relative mRNA expression levels of CD44, CD133, CXCR4, ALDH1, and CD24 are significantly reduced in miR-21 KO MiaPaCa-2 cells. The column graphic represents the average of three replicates of RNA isolated from MiaPaCa-2 control and its Kos. Data normalized according to RNA polymerase II (RPII) expression by fold analysis (n = 3, p < 0.05). p-values are indicated as * p ≤ 0.05; ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001; error bars indicate SD.
2.5. Flow Cytometry Analysis of PCSCs Expression in PDAC miR-21 KO Cells
Several key CSCs associated with PDAC, including CD24, CD133, and CD13 were assessed by flow cytometry in the miR-21 KO cells and compared with their expression in control cells. Results showed that CD133 was expressed at higher levels in Panc-1 control cells (96.9%; n = 3; Figure 5A), compared with Panc-1 KO2 (18.2%; 74.4-fold reduction; n = 3; p < 0.001; Figure 5B) and KO4 (21.8%; 69.3-fold reduction; n = 3; p < 0.001; Figure 5C,G). Similarly, in MiaPaCa-2 miR-21 Kos, the expression levels of CD133 were significantly higher in MiaPaCa-2 control (68.6%; n = 3; Figure 5D,H), compared with MiaPaCa-2 KO2 (17.9%; 52.3-fold decrease; n = 3; p < 0.001; Figure 5E,H) and KO4 (22.9%; 47.5-fold decrease; n = 3; p < 0.001; Figure 5F,H). Additionally, in the Panc-1 miR-21 Kos, the positive population for CD13 and CD24 presented a 100-fold and 95.7-fold reduction, respectively, in Panc-1 KO2 (0% and 4.3%, respectively; n = 3; Figure 6B) and 100-fold reduction in KO4 (0.2% and 0.1%, respectively; n = 3; Figure 6C), compared with what was observed in the Panc-1 control, where the positive population for both CD13 and CD24 was 97.4% (n = 3; Figure 6A). In the MiaPaCa-2 cell line, control cells were positive for CD24 (81.9%; n = 3; Figure 6D) and negative for CD13 (0%; n = 3; Figure 6D), whereas the percentage of negative cell population for both CD13 and CD24 PCSCs in the MiaPaCa-2 KO2 was 96.4% (90.1-fold; n = 3; Figure 6E) and 97.7% in KO4 (91.4-fold; n = 3; Figure 6F; Table 1). These results indicate that the depletion of miR-21 from both PDAC cell lines results in a decrease in the stemness markers CD133, CD24 and CD133.
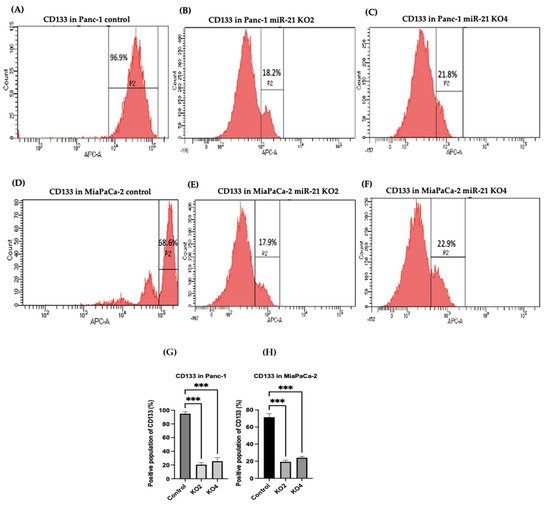
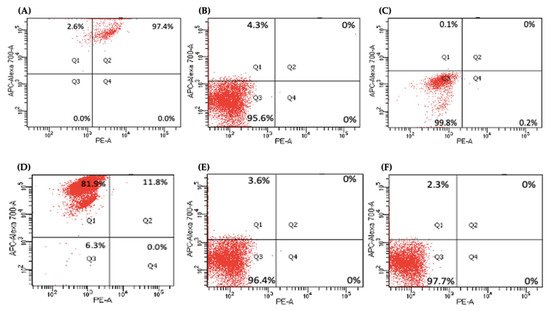

Figure 5. Flow cytometry analysis of CD133 in PDAC control and miR-21 KO cell lines. (A–C): Expression of CD133 in the Panc-1 miR-21 Kos and control: (A) Panc-1 vector alone (control cells); (B) miR-21 KO2; (C) miR-21 KO4. (D–F): Expression of CD133 in MiaPaCa-2 miR-21 Kos. (D) MiaPaCa-2 vector alone (control cells); € miR-21 KO2; (F) miR-21 KO4. (G) Percentage of the CD133 positive population in Panc-1 cells and their miR-21 KOs. CD133 expression was significantly increased in the Panc-1 vector alone compared with the miR-21 KOs in the Panc-1 cell line. The column graphic represents the average of three replicates of RNA isolated from Panc-1 control cells and their KOs. Data normalized according to RPII expression by fold analysis (n = 3, p < 0.05). (H) Percentage of the CD133 positive population in MiaPaCa-2 cells and their miR-21 KOs. CD133 expression was significantly increased in MiaPaCa-2 control cells compared with their miR-21 KOs. The column graphic represents the average of three replicates of RNA isolated from MiaPaCa-2 control cells and their KOs. Data normalized according to RPII expression by fold analysis (n = 3, p < 0.05). Numbers in the gated areas mark the percentages of cells that were positive for this specific marker. p-values are indicated as *** p ≤ 0.001; error bars indicate SD.

Figure 6. Flow cytometry analysis of CD24 and CD13 in miR-21 KO cell lines. (A–C) Expression of CD13 and CD24 in the Panc-1 KOs: (A) Panc-1 vector alone (control cells); (B) miR-21 KO2; (C) miR-21 KO4. Expression levels of both CD13 and CD24 were significantly reduced in Panc-1 miR-21 KO2 and KO4, compared with the Panc-1 control cells. (D–F): Expression of CD13 and CD24 in MiaPaCa-2 KOs: (D) MiaPaCa-2 vector alone (control cells); (E) miR-21 KO2; (F) miR-21 KO4. Expression levels of CD24 were significantly increased in MiaPaCa-2 control cells, while CD13 was significantly reduced not only in MiaPaCa-2 control cells but also in MiaPaCa-2 miR-21 KO2 and miR-21 KO4. APC-Alexa 700 symbolizes the CD24 marker, while PE stands for CD13. Numbers in the gated areas mark the percentages of cells that were positive for this specific marker.
Table 1. Percentage of expression of CD24 and CD13 in PDAC Cell Lines.
| Cancer Stem Cell Markers | Control | Panc-1 KO2 |
KO4 | Control | MiaPaCa-2 KO2 |
KO4 | |||
|---|---|---|---|---|---|---|---|---|---|
| CD24 | + | CD13 | + | 97.4% | 0% | 0% | 11.8% | 0% | 0% |
| CD24 | − | CD13 | − | 0% | 95.6% | 99.8% | 6.3% | 96.4% | 97.7% |
| CD24 | + | CD13 | − | 2.6% | 4.3% | 0.1% | 81.9% | 3.6% | 2.3% |
| CD24 | − | CD13 | + | 0% | 0% | 0.2% | 0% | 0% | 0% |
2.6. miR-21 KO Reduce Cellular Invasion in Panc-1 and MiaPaca-2 Cells
The invasiveness of both Panc-1 and MiaPaca-2 cells was studied by using Boyden chambers with Matrigel over a 16 h period. KO miR-21 resulted in a significant suppression of invasiveness by 31% and 22% in Panc-1; 27% and 16% in MiaPaca-2 cells (n = 3; p < 0.0001 and 0.001; p < 0.0001, respectively; Figure 7A). There was no significant change in the cell number over the 16 h in Panc-1 cells; however, a significant 3% reduction in MiaPaCa-2 cellular proliferation was detected in miR-21 KOs (n = 3; p > 0.05; p < 0.01, respectively; Figure 7B). The results show that miR-21 KO reduces invasiveness. The Panc-1 miR-21 KO cells did not exhibit a decrease in clonogenicity compared to control cells after 12 days, however, the MiaPaCa-2 miR-21 KO cells presented a reduction in proliferation compared to MiaPaCa-2 control (Figure 7C).
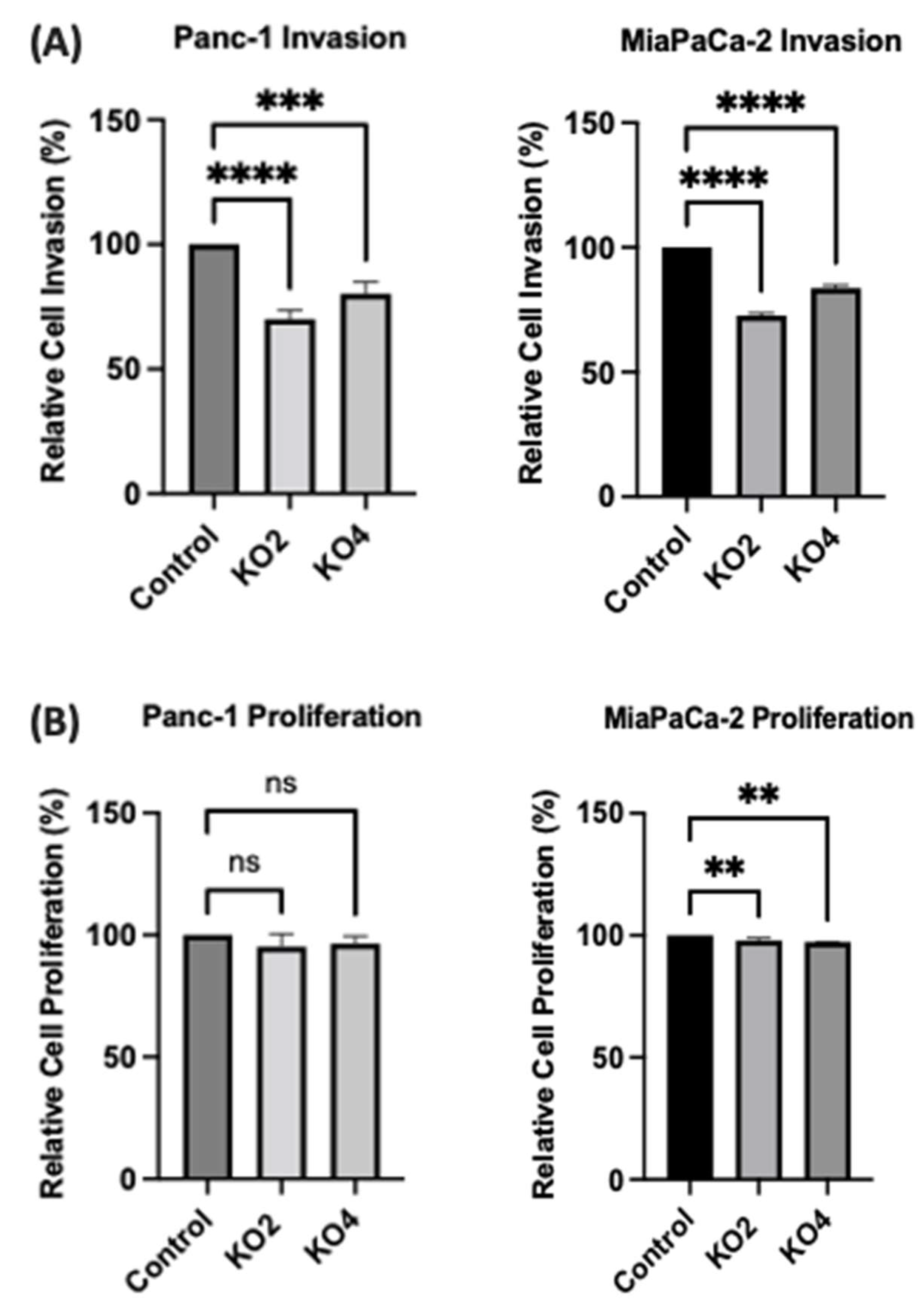
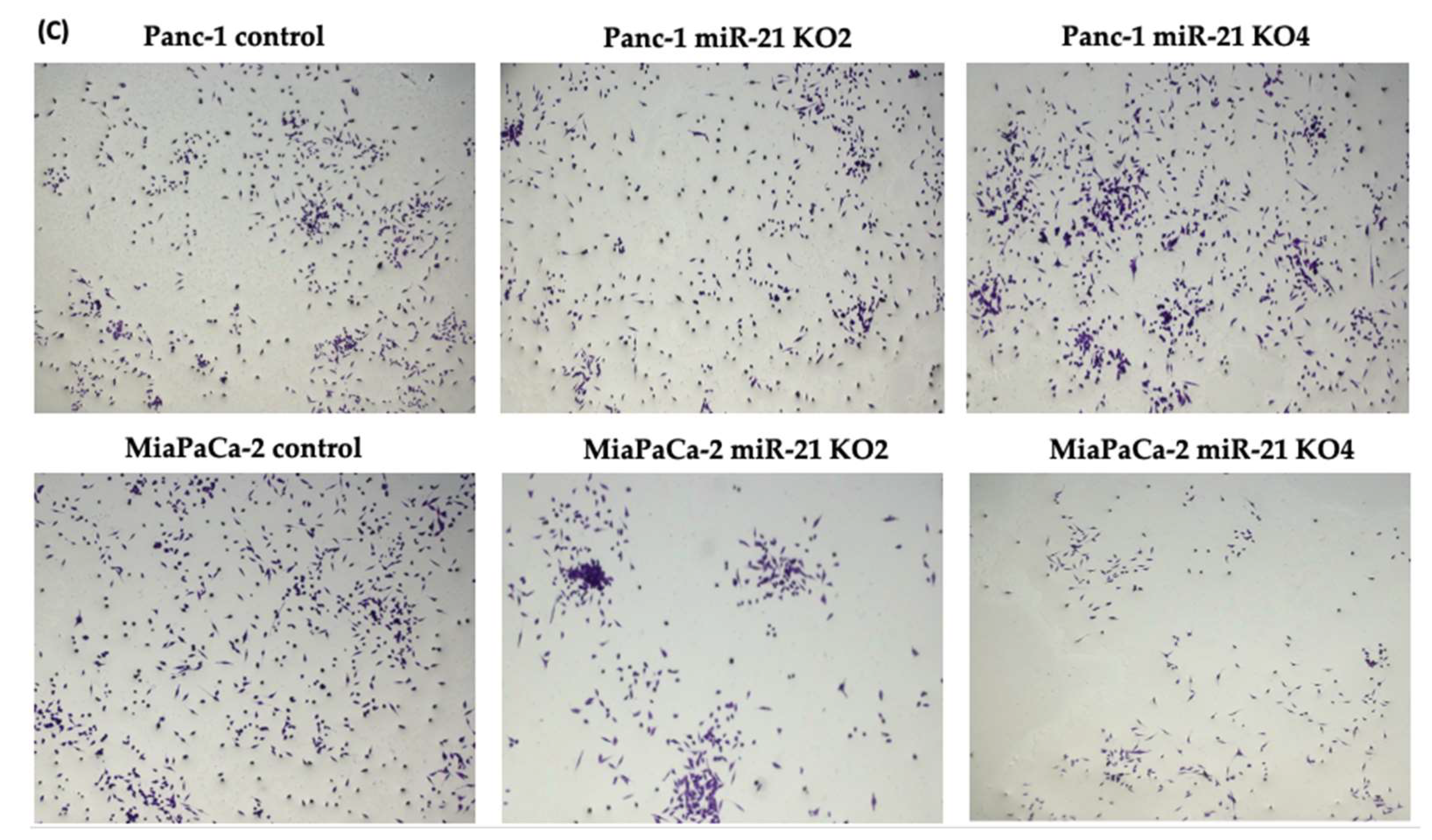
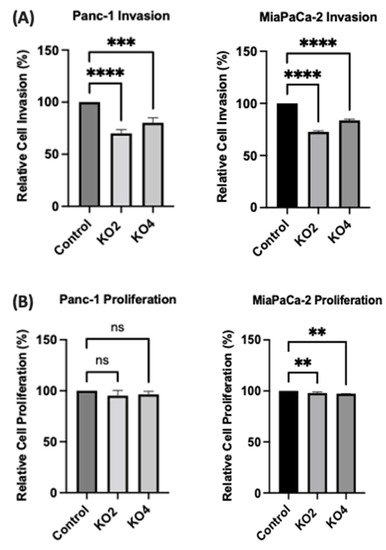
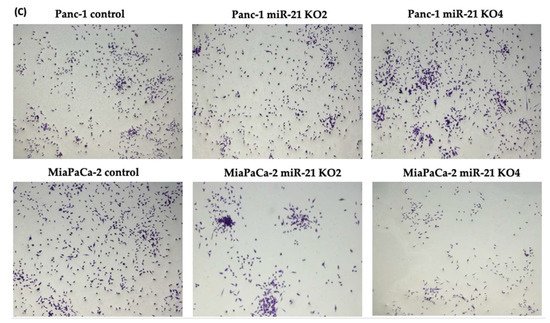




Figure 7. miR-21 reduced cellular invasion of Panc-1 and MiaPaca-2 cells. (A) Control and miR-21 KO cells were plated on Matrigel-coated transwell filters and the extent of invasion was determined after 16 h. The results are plotted as Relative Cell Invasion (%), which is the percentage of invaded cells compared to the total number of cells seeded. (B) The total cell number/proliferation did not change during the course of the experiment in the Panc-1 miR-21 KOs (n = 3; p > 0.05 for all), however, a small significant reduction was detected in the MiaPaCa-2 miR-21 KOs (n = 3; p < 0.01 for all). (C) Colony formation assay for Panc-1 and MiaPaCa-2 control and miR-21 KO cells. The colonies were observed with crystal violet staining of cells following 12 days. Images were obtained by using EVOS FL Auto 2 Imaging System (ThermoFisher, UK) with 10x magnification. p-values are indicated as ** p ≤ 0.01; *** p ≤ 0.001; **** p ≤ 0.0001; error bars indicate SD.
