Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 2 by Rita Xu and Version 1 by Yun Song.
The chloroplast has a central position in oxygenic photosynthesis and primary metabolism. In addition to these functions, the chloroplast has recently emerged as a pivotal regulator of plant responses to abiotic and biotic stress conditions. Chloroplasts have their own independent genomes and gene-expression machinery and synthesize phytohormones and a diverse range of secondary metabolites, a significant portion of which contribute the plant response to adverse conditions.
- chloroplast
- abiotic stress
- biotic stress
1. Introduction
As sessile organisms, plants have to cope with various environmental conditions that are unfavorable or stressful for their growth and development. These adverse environmental conditions include abiotic stresses such as drought, heat, chilling/freezing, salinity, and nutrient deficiency as well as biotic stresses such as herbivore attack and pathogen infection [1,2,3,4][1][2][3][4]. These abiotic and biotic stresses limit crop yields and cause tremendous economic losses. These adverse effects are getting worse due to climate change, increasing population, and less arable land [1,2][1][2]. Plants have evolved a number of strategies to sense stress signals and adapt to adverse environments. Increasing evidence has revealed the essential role of the chloroplast, an organelle for photosynthesis in green plants, in plant stress response and stress adaption [3,4,5][3][4][5].
The chloroplast is a double-membrane plant endosymbiotic organelle where photosynthesis takes place [5,6,7,8][5][6][7][8]. Chloroplasts have retained their own genomes, and the chloroplast genome contains approximately 120 genes involved in chloroplast activities such as energy production and gene expression [9,10][9][10]. Chloroplasts produce energy through photosynthesis and oxygen-release processes, which sustain plant growth and crop yield. As such, chloroplasts are responsible for the biosynthesis of active compounds such as amino acids, phytohormones, nucleotides, vitamins, lipids, and secondary metabolites [9]. Furthermore, the chloroplast plays a vital role in plant acclimation to environmental stresses [3,4,6][3][4][6]. When plants are in adverse environmental conditions, chloroplasts sense these stresses and synthesize biologically active compounds and phytohormones, which protect plants from environmental stresses. In addition, chloroplasts communicate with the nucleus through plastid-to-nucleus retrograde signaling to acclimate to environmental stresses (Figure 1) [3,11,12][3][11][12].
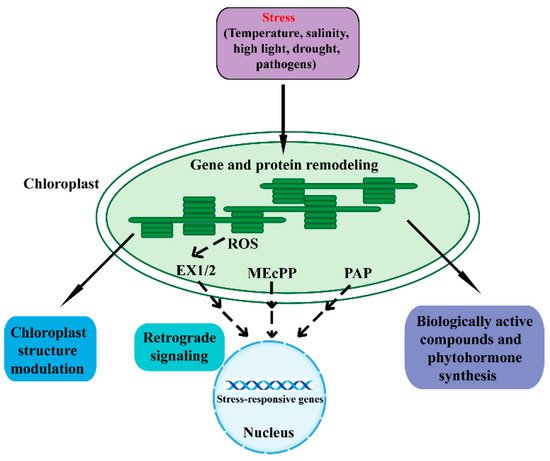
Figure 1. An overview of the various mechanisms in chloroplast response to adverse stresses. Adverse environmental stresses cause perturbations and generate signals in chloroplasts that regulate chloroplast gene expression and protein remodeling. A series of cellular activities are then triggered to restore chloroplast homeostasis. Adverse conditions can affect the structure, function, and development of chloroplasts. Chloroplasts synthesize biologically active compounds and phytohormones to acclimate to stresses. Moreover, the chloroplast is able to communicate its status to the nucleus through retrograde signaling to regulate nuclear stress-responsive genes. The SAL1/PAP, MEcPP, and ROS pathways act as important components of the chloroplast retrograde signaling pathway. Dashed lines indicate postulated regulation. ROS, reactive oxygen species; EX1/2, executor 1/2; MEcPP, methylerythritol cyclodiphosphate; PAP, phosphonucleotide 3′-phosphoadenosine 5′-phosphate.
Retrograde signaling pathways refer to the communications from the plastid to the nucleus [13]. The chloroplast functions as an environmental sensor. Fluctuations in the environment perturb chloroplast homeostasis, and the disturbance then drives the chloroplasts to communicate with the nucleus through retrograde signals. Eventually, plants remodel metabolism and gene expression to adapt to external stresses [7,11,12,13,14,15][7][11][12][13][14][15]. The retrograde signaling typically includes tetrapyrroles, phosphoadenosines, carotenoid oxidation products, isoprenoid precursors, carbohydrate metabolites, and reactive oxygen species (ROS) [16,17,18,19,20,21,22,23][16][17][18][19][20][21][22][23].
Under a series of unfavorable environmental conditions, chloroplasts generate the ROS retrograde signals including superoxide anion (O2−), hydrogen peroxide (H2O2), hydroxyl radical (OH▪), and singlet oxygen (1O2) [7,18,24,25,26,27][7][18][24][25][26][27]. The ROS in plants can severely threaten their health and viability. However, under stressful conditions, ROS function as a retrograde signal and modify the nuclear transcriptome to cope with these adverse stresses. Plants have evolved many strategies to maintain ROS dynamic equilibrium and normal photosynthetic efficiency, including complicated redox reaction chains and the ROS-scavenging system [18,26,27][18][26][27]. Singlet oxygen is generated at photosystem II (PSII) under high-light conditions through the excitation of ground-state triplet oxygen [7,11,24,28,29][7][11][24][28][29]. The singlet-oxygen-mediated transcriptional responses were first identified in the flu Arabidopsis mutant [30,31][30][31]. The functional FLU protein is a nuclear-encoded plastid protein that negatively regulates chlorophyll biosynthesis and consequently accumulates the strongly photosensitizing chlorophyll precursor protochlorophyllide. When dark-treated flu mutants are moved into the light, the mutants specifically accumulate singlet oxygen [32]. The singlet oxygen regulates a set of nuclear genes called singlet oxygen-responsive genes (SORGs), and most of these genes play a role in photosynthesis, carbon metabolism and plastid mRNA processing [7,33][7][33]. The activity of singlet oxygen signaling requires two nuclear-encoded proteins, Executor (Ex) 1 and 2, which are located in the thylakoid membrane of chloroplasts [31,32,34,35,36,37][31][32][34][35][36][37]. Furthermore, it has been reported that the plastid ATP-dependent zinc metalloprotease FtsH2 is involved in EX1/EX2 signaling [38,39][38][39]. Studies also find that β-cyclocitral is another singlet oxygen signaling pathway, which occurs independently of EX1 and EX2 [38,39,40][38][39][40]. Hydrogen peroxide acts as another retrograde signaling molecule. The specific role of hydrogen peroxide is identified through an RNAi line of thylakoid membrane-bound ascorbate peroxidase (tAPX) [41]. A number of putative components of the hydrogen peroxide signal transduction network were identified, including a series of transcription factors, mitogenactivated protein kinases (MAPKs) and miRNAs [7,42][7][42].
Methylerythritol cyclodiphosphate (MEcPP), an intermediate of the methylerythritol phosphate (MEP) pathway for plastid isoprenoid biosynthesis, functions as another retrograde signal to activate stress-responsive nuclear gene expression [23,43][23][43]. MEcPP was first identified by screening for mutants with elicited expression of hydroxyperoxide lyase (HPL). HPL is a nuclear stress-responsive gene and encodes a chloroplast enzyme, which plays a fundamental role in the defense-related molecules (such as the hormone jasmonic acid JA) synthesis [43]. The constitutively expressing HPL (ceh1) mutant was discovered. When exposed to high light or wounding stresses, plants accumulate MEcPP and regulate the expression of a series of nuclear genes [23,43,44,45][23][43][44][45]. Phosphonucleotide 3′-phosphoadenosine 5′-phosphate (PAP) is an important plastid metabolite and recently has emerged as a new retrograde signal for plant stress responses [16,46,47][16][46][47]. Chloroplast 3′ (2′), 5′-bisphosphate nucleotidase SAL1 can dephosphorylate PAP to adenosine monophosphate (AMP). When plants respond to drought and high light stresses, the activity of SAL1 is inhibited and PAP thus accumulates. PAP can regulate a series of nuclear stress-responsive genes [7]. Recent research has revealed the role of PAP in the drought stress response through regulating stomatal closure [46].
2. Chloroplast Response to Abiotic Stress
Advancing research has shown that chloroplasts play multifaceted roles in the plant response to various types of abiotic stress, including heat, chilling, salt, drought, and high light stresses. Here, we summarize the present state of knowledge on chloroplast responses to various abiotic stresses (Table 1).
Table 1. Summary of known chloroplast biological processes involved in plant stress response.
| Components | Species | Process/Stimulus | Molecular Function | Reference | |||||
|---|---|---|---|---|---|---|---|---|---|
| Heat stress | |||||||||
| SGR | Musa acuminata | Chlorophyll degradation | Stay-green protein | [56] | [48] | ||||
| Rca1 | Triticum aestivum | Modulate the activity of Rubisco | A catalytic chaperone | [57] | [49] | ||||
| CDJ2 | Solanum lycopersicum | , | Lycopersicon esculentum | Protect Rubisco activity | Chloroplast-targeted DnaJ protein | [58,59] | [50][51] | ||
| Ub2 | Triticum aestivum | Improve antioxidant capacity | Ubiquitin/26S proteasome system | [61] | [52] | ||||
| Hsp21 | Arabidopsis thaliana | Associate with the thylakoid membranes | Small heat-shock protein chaperone | [65] | [53] | ||||
| Chilling stress | |||||||||
| FBPase, SBPase | Zea mays | L. | Compensate for decreases in photosynthetic capacity | Enzymes involved in the Calvin cycle | [71] | [54] | |||
| DUA1 | Oryza sativa | Regulate chloroplast development | RNA-binding protein | [72] | [55] | ||||
| WHY1 | Solanum lycopersicum | Upregulate the PSII key D1 reaction center protein encoding gene | psbA | ; maintain high Rubisco content | WHIRLY proteins—the plant-specific DNA-binding proteins | [73,74] | [56][57] | ||
| CDJ1 | Lycopersicon esculentum | Maintain PSII activity | Chloroplast-targeted DnaJ protein | [75] | [58] | ||||
| RBD1 | Arabidopsis thaliana | Regulate chloroplast protein translation by influencing 23S rRNA processing | Chloroplast RNA-binding protein | [80] | [59] | ||||
| Salt stress | |||||||||
| SP1 | Arabidopsis thaliana | Induce the degradation of translocon at the outer envelope membrane of chloroplasts (TOC) | Uiquitin E3 ligase suppressor of PPI1 locus 1 | [83,84] | [60][61] | ||||
| Hsp70, Clp protease |
Arabidopsis thaliana | Protect plants from salinity-triggered oxidative stress | Hsp70—chaperone; Clp—a protease system |
[85] | [62] | ||||
| RH22 | Brassica rapa | Affect chloroplast gene translation | Cloroplast-targeted DEAD-box RNA helicase | [86] | [63] | ||||
| CspA, CspB | Oryza sativa | Impart SGR phenotype | RNA-binding bacterial chaperones | [87] | [64] | ||||
| High-light stress | |||||||||
| SAL1-PAP | Arabidopsis thaliana | Regulate stress-inducible nuclear genes | Components of retrograde pathway | [16] | |||||
| FtSH | Chlamydomonas reinhardtii | Control the quality of thylakoid membrane proteins | Thylakoid membrane protease | [91] | [65] | ||||
| Drought stress | |||||||||
| SAL1-PAP | Arabidopsis thaliana | Regulate the stress-inducible nuclear genes | Components of retrograde pathway | [16] | |||||
| RH22 | Brassica rapa | Affect chloroplast genes’ translation | Chloroplast-targeted DEAD-box RNA helicase | [86] | [63] | ||||
| CTR1 | Oryza sativa | Interact with two chloroplast-localized proteins, OsCP12 and OsRP1 | RING Ub E3 ligase | [93] | [66] | ||||
| PhyB | Oryza sativa | Repress the activity of ascorbate peroxidase | Phytochrome B | [94] | [67] | ||||
| LOX6 | Zea mays | Additional storage of nitrogen | Mesophyll lipoxygenase in chloroplast | [95] | [68] | ||||
| NADP-ME4 | Salsola laricifolia | Alleviate chlorophyll content decrease and PSII photochemical efficiency | NADP-malic enzyme | [96] | [69] | ||||
| BBX21 | Solanum tuberosum | Reduce chloroplast electron transport capacity | B-box (BBX) protein | [97] | [70] | ||||
| Biotic stress | |||||||||
| PP2C62, PP2C26 | Arabidopsis thaliana | Catalyze the dephosphorylation of the photosynthesis-related protein, chaperonin-60 | Components of the serine/threonine-specific protein phosphatase family | [107] | [71] | ||||
| XopL | Nicotiana benthamiana | Eliminate stromules formation and chloroplast relocation | E3 ubiquitin ligase | [108] | [72] | ||||
| WKS1 | Triticum aestivum | Phosphorylate thylakoid-associated ascorbate peroxidase (tAPX) and detoxify peroxides; phosphorylate an extrinsic member of photosystem II (PSII) PsbO | A serine/threonine kinase | [109,110] | [73][74] | ||||
| THF1 | Nicotiana benthamiana | Maintain chloroplast homeostasis | Chloroplastic protein thylakoid formation1 | [111] | [75] | ||||
| APX8 | Oryza sativa | Regulate H | 2 | O | 2 | accumulation | Thylakoid membrane-bound ascorbate peroxidase | [112] | [76] |
| LHCB5 | Oryza sativa | Regulate ROS accumulation | Light-harvesting complex II protein | [113] | [77] | ||||
| NTRC | Arabidopsis thaliana | Modulate chloroplast-generated ROS | Redox detoxification system NADPH-dependent thioredoxin reductase C | [114] | [78] | ||||
| MPK3, MPK6 | Arabidopsis thaliana | Manipulate plant photosynthetic activities and promote ROS accumulation | Mitogen-activated protein kinase | [115] | [79] | ||||
| PsbQ | Nicotiana benthamiana, Arabidopsis thaliana | A target for pathogen suppression and contributes to plant immunity responses | The oxygen evolving complex of photosystem II | [118] | [80] | ||||
| ALC | Solanum lycopersicum | , | Arabidopsis thaliana | Regulate disease-associated necrotic cell death | Chloroplast genes with altered responses to coronatine | [119] | [81] | ||
| RipAL | Ralstonia solanacearum | Localized to chloroplasts and targeted chloroplast lipids | Type III effector proteins | [121] | [82] | ||||
| Tsn1 | Triticum aestivum | Interact with effector protein ToxA | A unique wheat disease resistance-like gene, regulated by the circadian clock and light | [122] | [83] | ||||
| Rpi-vnt1.1 | Nicotiana benthamiana | Recognize the effector protein AVRvnt1, and mediate a light-dependent immune response | A nucleotide-binding leucine-rich repeat (NLR) protein | [123] | [84] | ||||
| CAS | Arabidopsis thaliana | Recognize the effector protein, and interfere with the salicylic acid (SA) signaling pathway | A chloroplast-localized calcium-sensing receptor | [124] | [85] | ||||
2.1. Response to Heat Stress
Leaf photosynthesis is substantially affected by abnormal temperature stresses, including heat stress which is usually 10–15 °C above an optimum temperature for plant growth or chilling stress which occurs in the temperatures range 0–15 °C [27,48][27][86]. Chloroplasts play an essential role in activation of physiological adaptive processes to these adverse temperature stresses.
It is reported that the chloroplast is sensitive to high-temperature stress during photosynthesis [48][86]. Research has revealed a close association between the chloroplast-related genes and high-temperature stress in the model plant rice. The expression of more than two hundred genes was upregulated in response to heat stress [49][87]. In a series of plant species, heat-induced leaf chlorosis has been observed [48][86]. After heat treatment, the activities of chlorophyll-degrading enzymes increased significantly, and the activity of key chlorophyll-synthesizing enzymes was unchanged [50][88]. Studies on genetic variations in hybrids of colonial (Agrostis capillaris) x creeping bentgrass (Agrostis stolonifera) indicated that the rapid breakdown of chlorophyll induced by heat stress was related to activation of genes encoding chlorophyllase and pheophytinase, and pheophytinase (PPH) activity [51][89]. The stay-green physiological traits were selected to evaluate the heat tolerance of plants and reveal the potential mechanism of heat damage associated with alterations in Chl metabolism and antioxidant and photosynthetic capacity. In creeping bentgrass species, adaptability to high temperature and stay-green genotypes was highly associated with chlorophyll metabolism [52][90]. Stay-green (SGR) genes encode magnesium dechelatase, and are involved in chlorophyll (Chl) degradation. Stay-green (SGR) homologs remove magnesium from Chl a, which is one of the most important components in the Chl degradation pathway in plants [53,54,55][91][92][93]. Under high-temperature conditions, accumulation of sugars in the peel was induced in bananas, and these sugars regulated Chl degradation through SGR proteins [56][48].
A series of research has shown that regulation of the activity of photosynthetic pathway enzyme Rubisco contributes to plant adaption to the heat stress. Rubisco activase (Rca1), a catalytic chaperone involved in modulating the Rubisco activity, plays a role in wheat response to heat stress [57][49]. The tomato (Solanum lycopersicum) chloroplast-targeted DnaJ protein (SlCDJ2) is located in the thylakoids and stroma of the chloroplasts. When plants respond to heat stress, SlCDJ2 protects Rubisco activity, and contributes to maintenance of CO2 assimilation capacity, which establishes a role for SlCDJ2 in coping with heat stress [58][50]. The tomato (Lycopersicon esculentum) chloroplast-targeted DnaJ protein (LeCDJ1) also plays a role in plant response to high temperature [59][51].
Furthermore, the activity of PSI and PSII is severely affected by high-temperature conditions. In A. thaliana, heat stress can modulate the transcript accumulation of the plastid-encoded PSI and PSII genes such as the psaA, psaB, psbA, psbD, and psbN. Furthermore, the regulation is performed in part via the expression of HS-responsive nuclear genes for the plastid transcription machinery [60][94]. In wheat plants, the increased photosynthetic rate, improved ATPase activity in the thylakoid membrane, and enhanced efficiency of PSII photochemistry, which was achieved by overexpression of the ubiquitin/26S proteasome system TaUb2, contributed to plants coping with high-temperature stress [61][52]. When plants are exposed to transient heat waves, insufficient PSI photoprotection including the regulation of linear electron transport and the prevention of over-reduction, may affect the wheat photosynthetic capacity and make plants more susceptible to heat stress [62][95].
When plants were subjected to high-temperature stress, they generated large amounts of ROS and initiated related signaling events to survive the adverse environments [63][96]. In a recent study, researchers found that, under heat stress, the ROS levels contributed to mitigation of PSII photoinhibition in a coffee crop [64][97]. When plants respond to heat stress, the chloroplast heat-shock protein (Hsp) 21 becomes associated with the thylakoid membranes and plays a role in plant stress resistance [65][53].
2.2. Response to Low-Temperature Stress
In addition to high-temperature tolerance, chloroplasts are involved in plant cold-stress response. A series of studies have reported that chloroplasts can also perceive chilling stress signals, promote photosynthesis, and enhance plant resistance to adverse environment stress [27]. Previous and recent studies showed that low-temperature conditions could affect the abundance of various proteins involved in photosynthesis [27]. Through iTRAQ-based proteomic analysis between chilling-tolerant and chilling-sensitive rice lines, scientists revealed the dynamic response of chloroplast photosynthetic proteins under chilling conditions [66][98]. Systematic analysis of cold-stress response using transcriptome data was performed in rice, and the stay-green (SGR) proteins were identified as hub genes in this life process. Furthermore, SGR proteins were involved in the crosstalk between cold-stress responses and diurnal rhythmic patterns, providing new insights in understanding the plant environmental stress response against climate change [67][99].
Chilling can affect the structure, function, and development of chloroplasts [68][100]. Scientists revealed that chilling can induce structural changes during chloroplast biogenesis in cucumber cotyledons [69][101]. When exposed to low temperatures, plant chloroplasts changed the content of unsaturated fatty acids in chloroplast membranes to increase plant tolerance to the adverse temperatures [70][102]. When plants adapted to chilling stresses, the activities of chloroplast dark-reaction-related enzyme were also regulated. For instance, the reductive activity of two key enzymes involved in the Calvin cycle, fructose-1,6-diphosphatase (FBPase) and isoheptanone-1,7-diphosphatase (SBPase) was significantly reduced [71][54]. Low-temperature stress hinders chloroplast development and plant photosynthesis. The rice (Oryza sativa) RNA-binding protein DUA1 is required for RNA editing of the rps8-182 site, and plays a vital role in chloroplast development under low-temperature conditions [72][55]. The chloroplast gene psbA encodes the key D1 reaction center protein of PSII. Recently, the tomato (Solanum lycopersicum) WHIRLY1 (SlWHY1) was found to be induced by chilling conditions and could upregulate the transcription level of psbA through directly binding to the upstream region of its promoter (the sequence “GTTACCCT”). The increased D1 abundance enhanced plant resistance to photoinhibition caused by chilling stresses [73][56]. Scientists also found that SlWHY1 could increase the expression level of RbcS1, a member of the Rubisco small-subunit (RBCS) multigene family, and help plants maintain high Rubisco content under low-temperature conditions [74][57]. Despite the role of chloroplast-targeted DnaJ protein in plant response to heat stress, the function of tomato (Lycopersicon esculentum) chloroplast-targeted DnaJ protein (LeCDJ1) under chilling stress was also investigated. LeCDJ1 showed essential functions in maintaining PSII activity under low-temperature stress [75][58].
The ROS function has an important retrograde signal in chloroplasts under a series of unfavorable environmental conditions. Chilling stress can regulate the redox state of chloroplasts. When plants respond to chilling stress, the photosynthetic electron transport chain in chloroplasts transfers excess electrons to O2 and causes O2- increase in bermudagrass [76][103]. In A. thaliana, regulation of chloroplast-to-nucleus ROS signaling is a strategy to promote plants acclimation to cold stress [77][104]. Researchers revealed that melatonin increases the chilling tolerance of cucumber seedlings through regulating the ROS balance [78][105]. Scientists have also found that, under chilling stress, exogenous applications of acetylsalicylic acid can enhance the chloroplast antioxidant system activity and thus improve the tolerance of plants under low temperatures [79][106].
References
- Zhu, J.K. Abiotic Stress Signaling and Responses in Plants. Cell 2016, 167, 313–324.
- Fedoroff, N.V.; Battisti, D.S.; Beachy, R.N.; Cooper, P.J.; Fischhoff, D.A.; Hodges, C.N.; Knauf, V.C.; Lobell, D.; Mazur, B.J.; Molden, D.; et al. Radically Rethinking Agriculture for the 21st Century. Science 2010, 327, 833–834.
- Zhang, Y.; Zhang, A.; Li, X.; Lu, C. The Role of Chloroplast Gene Expression in Plant Responses to Environmental Stress. Int. J. Mol. Sci. 2020, 21, 6082.
- Littlejohn, G.R.; Breen, S.; Smirnoff, N.; Grant, M. Chloroplast Immunity Illuminated. New Phytol. 2021, 229, 3088–3107.
- Serrano, I.; Audran, C.; Rivas, S. Chloroplasts at Work During Plant Innate Immunity. J. Exp. Bot. 2016, 67, 3845–3854.
- Mamaeva, A.; Taliansky, M.; Filippova, A.; Love, A.J.; Golub, N.; Fesenko, I. The Role of Chloroplast Protein Remodeling in Stress Responses and Shaping of The Plant Peptidome. New Phytol. 2020, 227, 1326–1334.
- Watson, S.J.; Sowden, R.G.; Jarvis, P. Abiotic Stress-induced Chloroplast Proteome Remodelling: A Mechanistic Overview. J. Exp. Bot. 2018, 69, 2773–2781.
- Bose, J.; Munns, R.; Shabala, S.; Gilliham, M.; Pogson, B.; Tyerman, S.D. Chloroplast Function and Ion Regulation in Plants Growing on Saline Soils: Lessons from Halophytes. J. Exp. Bot. 2017, 68, 3129–3143.
- Daniell, H.; Lin, C.S.; Yu, M.; Chang, W.J. Chloroplast Genomes: Diversity, Evolution, and Applications in Genetic Engineering. Genome Biol. 2016, 17, 134.
- Huo, Y.; Gao, L.; Liu, B.; Yang, Y.; Kong, S.; Sun, Y.; Yang, Y.; Wu, X. Complete Chloroplast Genome Sequences of Four Allium Species: Comparative and Phylogenetic Analyses. Sci. Rep. 2019, 9, 12250.
- Chan, K.X.; Phua, S.Y.; Crisp, P.; McQuinn, R.; Pogson, B.J. Learning the Languages of the Chloroplast: Retrograde Signaling and Beyond. Annu. Rev. Plant Biol. 2016, 67, 25–53.
- Crawford, T.; Lehotai, N.; Strand, Å. The Role of Retrograde Signals during Plant Stress Responses. J. Exp. Bot. 2018, 69, 2783–2795.
- de Souza, A.; Wang, J.Z.; Dehesh, K. Retrograde Signals: Integrators of Interorganellar Communication and Orchestrators of Plant Development. Annu. Rev. Plant Biol. 2017, 6, 85–108.
- Wu, G.Z.; Meyer, E.H.; Wu, S.; Bock, R. Extensive Posttranscriptional Regulation of Nuclear Gene Expression by Plastid Retrograde Signals. Plant Physiol. 2019, 180, 2034–2048.
- Fernandez, J.C.; Burch-Smith, T.M. Chloroplasts as Mediators of Plant Biotic Interactions over Short and Long Distances. Curr. Opin. Plant Biol. 2019, 50, 148–155.
- Estavillo, G.M.; Crisp, P.A.; Pornsiriwong, W.; Wirtz, M.; Collinge, D.; Carrie, C.; Giraud, E.; Whelan, J.; David, P.; Javot, H.; et al. Evidence for A SAL1-PAP Chloroplast Retrograde Pathway that Functions in Drought and High Light Signaling in Arabidopsis. Plant Cell 2011, 23, 3992–4012.
- Häusler, R.E.; Heinrichs, L.; Schmitz, J.; Flügge, U.I. How Sugars might Coordinate Chloroplast and Nuclear Gene Expression during Acclimation to High Light Intensities. Mol. Plant 2014, 7, 1121–1137.
- Ramel, F.; Birtic, S.; Ginies, C.; Soubigou-Taconnat, L.; Triantaphylidès, C.; Havaux, M. Carotenoid Oxidation Products are Stress Signals that Mediate Gene Responses to Singlet Oxygen in Plants. Proc. Natl. Acad. Sci. USA 2012, 109, 5535–5540.
- Shumbe, L.; Bott, R.; Havaux, M. Dihydroactinidiolide, a High Light-Induced β-carotene Derivative that Can Regulate Gene Expression and Photoacclimation in Arabidopsis. Mol. Plant 2014, 7, 1248–1251.
- Strand, A.; Asami, T.; Alonso, J.; Ecker, J.R.; Chory, J. Chloroplast to Nucleus Communication Triggered by Accumulation of Mg-protoporphyrinIX. Nature 2003, 421, 79–83.
- Vogel, M.O.; Moore, M.; König, K.; Pecher, P.; Alsharafa, K.; Lee, J.; Dietz, K.J. Fast Retrograde Signaling in Response to High Light Involves Metabolite Export, MITOGEN-ACTIVATED PROTEIN KINASE6, and AP2/ERF Transcription Factors in Arabidopsis. Plant Cell 2014, 26, 1151–1165.
- Woodson, J.D.; Perez-Ruiz, J.M.; Schmitz, R.J.; Ecker, J.R.; Chory, J. Sigma Factor-Mediated Plastid Retrograde Signals Control Nuclear Gene Expression. Plant J. 2013, 73, 1–13.
- Xiao, Y.; Savchenko, T.; Baidoo, E.E.; Chehab, W.E.; Hayden, D.M.; Tolstikov, V.; Corwin, J.A.; Kliebenstein, D.J.; Keasling, J.D.; Dehesh, K. Retrograde Signaling by the Plastidial Metabolite MEcPP Regulates Expression of Nuclear Stress-response Genes. Cell 2012, 149, 1525–1535.
- Mignolet-Spruyt, L.; Xu, E.; Idänheimo, N.; Hoeberichts, F.A.; Mühlenbock, P.; Brosché, M.; Van Breusegem, F.; Kangasjärvi, J. Spreading the News: Subcellular and Organellar Reactive Oxygen Species Production and Signalling. J. Exp. Bot. 2016, 67, 3831–3844.
- Tripathy, B.C.; Oelmüller, R. Reactive Oxygen Species Generation and Signaling in Plants. Plant Signal Behav. 2012, 7, 1621–1633.
- Chan, K.X.; Crisp, P.A.; Estavillo, G.M.; Pogson, B.J. Chloroplast-to-Nucleus Communication: Current Knowledge, Experimental Strategies and Relationship to Drought Stress Signaling. Plant Signal Behav. 2010, 5, 1575–1582.
- Gan, P.; Liu, F.; Li, R.; Wang, S.; Luo, J. Chloroplasts- Beyond Energy Capture and Carbon Fixation: Tuning of Photosynthesis in Response to Chilling Stress. Int. J. Mol. Sci. 2019, 20, 5046.
- Triantaphylidès, C.; Havaux, M. Singlet Oxygen in Plants: Production, Detoxification and Signaling. Trends Plant Sci. 2009, 14, 219–228.
- González-Pérez, S.; Gutiérrez, J.; García-García, F.; Osuna, D.; Dopazo, J.; Lorenzo, Ó.; Revuelta, J.L.; Arellano, J.B. Early Transcriptional Defense Responses in Arabidopsis Cell Suspension Culture under High-Light Conditions. Plant Physiol. 2011, 156, 1439–1456.
- Meskauskiene, R.; Nater, M.; Goslings, D.; Kessler, F.; Op den Camp, R.; Apel, K.F.L.U. A Negative Regulator of Chlorophyll Biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2001, 98, 12826–12831.
- Wagner, D.; Przybyla, D.; Op den Camp, R.; Kim, C.; Landgraf, F.; Lee, K.P.; Würsch, M.; Laloi, C.; Nater, M.; Hideg, E.; et al. The Genetic Basis of Singlet Oxygen-Induced Stress Responses of Arabidopsis thaliana. Science 2004, 306, 1183–1185.
- Op den Camp, R.G.; Przybyla, D.; Ochsenbein, C.; Laloi, C.; Kim, C.; Danon, A.; Wagner, D.; Hideg, E.; Göbel, C.; Feussner, I.; et al. Rapid Induction of Distinct Stress Responses After the Release of Singlet Oxygen in Arabidopsis. Plant Cell 2003, 15, 2320–2332.
- Page, M.T.; McCormac, A.C.; Smith, A.G.; Terry, M.J. Singlet Oxygen Initiates a Plastid Signal Controlling Photosynthetic Gene Expression. New Phytol. 2017, 213, 1168–1180.
- Lee, K.P.; Kim, C.; Landgraf, F.; Apel, K. EXECUTER1- and EXECUTER2-Dependent Transfer of Stress-Related Signals from the Plastid to the Nucleus of Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2007, 104, 10270–10275.
- Kim, C.; Apel, K. Singlet Oxygen-Mediated Signaling in Plants: Moving from Flu to Wild Type Reveals an Increasing Complexity. Photosynth. Res. 2013, 116, 455–464.
- Uberegui, E.; Hall, M.; Lorenzo, Ó.; Schröder, W.P.; Balsera, M. An Arabidopsis Soluble Chloroplast Proteomic Analysis Reveals the Participation of the Executer Pathway in Response to Increased Light Conditions. J. Exp. Bot. 2015, 66, 2067–2077.
- Carmody, M.; Crisp, P.A.; d’Alessandro, S.; Ganguly, D.; Gordon, M.; Havaux, M.; Albrecht-Borth, V.; Pogson, B.J. Uncoupling High Light Responses from Singlet Oxygen Retrograde Signaling and Spatial-Temporal Systemic Acquired Acclimation. Plant Physiol. 2016, 171, 1734–1749.
- Wang, L.; Kim, C.; Xu, X.; Piskurewicz, U.; Dogra, V.; Singh, S.; Mahler, H.; Apel, K. Singlet Oxygen- and EXECUTER1-Mediated Signaling Is Initiated in Grana Margins and Depends on the Protease FtsH2. Proc. Natl. Acad. Sci. USA 2016, 113, E3792–E3800.
- Dogra, V.; Duan, J.; Lee, K.P.; Lv, S.; Liu, R.; Kim, C. FtsH2-Dependent Proteolysis of EXECUTER1 Is Essential in Mediating Singlet Oxygen-Triggered Retrograde Signaling in Arabidopsis thaliana. Front Plant Sci. 2017, 8, 1145.
- Shumbe, L.; D’Alessandro, S.; Shao, N.; Chevalier, A.; Ksas, B.; Bock, R.; Havaux, M. METHYLENE BLUE SENSITIVITY 1 (MBS1) is Required for Acclimation of Arabidopsis to Singlet Oxygen and Acts Downstream of β-cyclocitral. Plant Cell Environ. 2017, 40, 216–226.
- Maruta, T.; Noshi, M.; Tanouchi, A.; Tamoi, M.; Yabuta, Y.; Yoshimura, K.; Ishikawa, T.; Shigeoka, S. H2O2-Triggered Retrograde Signalingfrom Chloroplasts to Nucleus Plays Specific Role in Response to Stress. J. Biol. Chem. 2012, 287, 11717–11729.
- Petrov, V.D.; Van Breusegem, F. Hydrogen Peroxide-A Central Hub for Information Flow in Plant Cells. AoB Plants 2012, 2012, pls014.
- Benn, G.; Bjornson, M.; Ke, H.; De Souza, A.; Balmond, E.I.; Shaw, J.T.; Dehesh, K. Plastidial Metabolite MEcPP Induces a Transcriptionally Centered Stress-response Hub Via the Transcription Factor CAMTA3. Proc. Natl. Acad. Sci. USA 2016, 113, 8855–8860.
- Walley, J.; Xiao, Y.; Wang, J.Z.; Baidoo, E.E.; Keasling, J.D.; Shen, Z.; Briggs, S.P.; Dehesh, K. Plastid-Produced Interorgannellar Stress Signal MEcPP Potentiates Induction of the Unfolded Protein Response in Endoplasmic Reticulum. Proc. Natl. Acad. Sci. USA 2015, 112, 6212–6217.
- Lemos, M.; Xiao, Y.; Bjornson, M.; Wang, J.Z.; Hicks, D.; Souza, A.; Wang, C.Q.; Yang, P.; Ma, S.; Dinesh-Kumar, S.; et al. The Plastidial Retrograde Signal Methyl Erythritol Cyclopyrophosphate Is a Regulator of Salicylic Acid and Jasmonic Acid Crosstalk. J. Exp. Bot. 2016, 67, 1557–1566.
- Pornsiriwong, W.; Estavillo, G.M.; Chan, K.X.; Tee, E.E.; Ganguly, D.; Crisp, P.A.; Phua, S.Y.; Zhao, C.; Qiu, J.; Park, J.; et al. A Chloroplast Retrograde Signal, 3’-Phosphoadenosine 5’-Phosphate, Acts as A Secondary Messenger in Abscisic Acid Signaling in Stomatal Closure and Germination. eLife 2017, 6, e23361.
- Chan, K.X.; Mabbitt, P.D.; Phua, S.Y.; Mueller, J.W.; Nisar, N.; Gigolashvili, T.; Stroeher, E.; Grassl, J.; Arlt, W.; Estavillo, G.M.; et al. Sensing and Signaling of Oxidative Stress in Chloroplasts by Inactivation of the SAL1 Phosphoadenosine Phosphatase. Proc. Natl. Acad. Sci. USA 2016, 113, E4567–E4576.
- Yang, X.; Pang, X.; Xu, L.; Fang, R.; Huang, X.; Guan, P.; Lu, W.; Zhang, Z. Accumulation of Soluble Sugars in Peel at High Temperature Leads to Stay-green Ripe Banana Fruit. J. Exp. Bot. 2009, 60, 4051–4062.
- Kumar, R.R.; Goswami, S.; Singh, K.; Dubey, K.; Singh, S.; Sharma, R.; Verma, N.; Kala, Y.K.; Rai, G.K.; Grover, M.; et al. Identification of Putative RuBisCo Activase (TaRca1)-The Catalytic Chaperone Regulating Carbon Assimilatory Pathway in Wheat (Triticum aestivum) under the Heat Stress. Front. Plant Sci. 2016, 7, 986.
- Wang, G.; Kong, F.; Zhang, S.; Meng, X.; Wang, Y.; Meng, Q. A Tomato Chloroplast-Targeted DnaJ Protein Protects Rubisco Activity under Heat Stress. J. Exp. Bot. 2015, 66, 3027–3040.
- Kong, F.; Deng, Y.; Wang, G.; Wang, J.; Liang, X.; Meng, Q. LeCDJ1, A Chloroplast DnaJ Protein, Facilitates Heat Tolerance in Transgenic Tomatoes. J. Integr. Plant Biol. 2014, 56, 63–74.
- Tian, F.; Gong, J.; Zhang, J.; Feng, Y.; Wang, G.; Guo, Q.; Wang, W. Overexpression of Monoubiquitin Improves Photosynthesis in Transgenic Tobacco Plants Following High Temperature Stress. Plant Sci. 2014, 226, 92–100.
- Bernfur, K.; Rutsdottir, G.; Emanuelsson, C. The Chloroplast-Localized Small Heat Shock Protein Hsp21 Associates with the Thylakoid Membranes in Heat-Stressed Plants. Protein Sci. 2017, 26, 1773–1784.
- Kingston-Smith, A.H.; Harbinson, J.; Williams, J.; Foyer, C.H. Effect of Chilling on Carbon Assimilation, Enzyme Activation, and Photosynthetic Electron Transport in the Absence of Photoinhibition in Maize Leaves. Plant Physiol. 1997, 114, 1039–1046.
- Cui, X.; Wang, Y.; Wu, J.; Han, X.; Gu, X.; Lu, T.; Zhang, Z. The RNA Editing Factor DUA1 is Crucial to Chloroplast Development at Low Temperature in Rice. New Phytol. 2019, 221, 834–849.
- Zhuang, K.; Kong, F.; Zhang, S.; Meng, C.; Yang, M.; Liu, Z.; Wang, Y.; Ma, N.; Meng, Q. Whirly1 Enhances Tolerance to Chilling Stress in Tomato via Protection of Photosystem II and Regulation of Starch Degradation. New Phytol. 2019, 221, 1998–2012.
- Zhuang, K.; Wang, J.; Jiao, B.; Chen, C.; Zhang, J.; Ma, N.; Meng, Q. WHIRLY1 Maintains Leaf Photosynthetic Capacity in Tomato by Regulating the Expression of RbcS1 Under Chilling Stress. J. Exp. Bot. 2020, 71, 3653–3663.
- Kong, F.; Deng, Y.; Zhou, B.; Wang, G.; Wang, Y.; Meng, Q. A Chloroplast-Targeted DnaJ Protein Contributes to Maintenance of Photosystem II under Chilling Stress. J. Exp. Bot. 2014, 65, 143–158.
- Wang, S.; Bai, G.; Wang, S.; Yang, L.; Yang, F.; Wang, Y.; Zhu, J.K.; Hua, J. Chloroplast RNA-Binding Protein RBD1 Promotes Chilling Tolerance through 23S rRNA Processing in Arabidopsis. PLoS Genet. 2016, 12, e1006027.
- Ling, Q.; Jarvis, P. Regulation of Chloroplast Protein Import by the Ubiquitin E3 Ligase SP1 Is Important for Stress Tolerance in Plants. Curr. Biol. 2015, 25, 2527–2534.
- Ling, Q.; Jarvis, P. Plant Signaling: Ubiquitin Pulls the Trigger on Chloroplast Degradation. Curr. Biol. 2016, 26, R38–R40.
- Pulido, P.; Llamas, E.; Rodriguez-Concepcion, M. Both Hsp70 Chaperone and Clp Protease Plastidial Systems are Required for Protection Against Oxidative Stress. Plant Signal Behav. 2017, 12, e1290039.
- Nawaz, G.; Lee, K.; Park, S.J.; Kim, Y.O.; Kang, H. A Chloroplast-Targeted Cabbage DEAD-box RNA Helicase BrRH22 Confers Abiotic Stress Tolerance to Transgenic Arabidopsis Plants by Affecting Translation of Chloroplast Transcripts. Plant Physiol. Biochem. 2018, 127, 336–342.
- Guddimalli, R.; Somanaboina, A.K.; Palle, S.R.; Edupuganti, S.; Kummari, D.; Palakolanu, S.R.; Naravula, J.; Gandra, J.; Qureshi, I.A.; Marka, N.; et al. Overexpression of RNA-binding Bacterial Chaperones in Rice Leads to Stay-green Phenotype, Improved Yield and Tolerance to Salt and Drought Stresses. Physiol. Plant. 2021, 173, 1351–1368.
- Malnoë, A.; Wang, F.; Girard-Bascou, J.; Wollman, F.A.; de Vitry, C. Thylakoid FtsH Protease Contributes to Photosystem II and Cytochrome B6f Remodeling in Chlamydomonas reinhardtii under Stress Conditions. Plant Cell 2014, 26, 373–390.
- Lim, S.D.; Lee, C.; Jang, C.S. The Rice RING E3 ligase, OsCTR1, Inhibits Trafficking to The Chloroplasts of OsCP12 and OsRP1, and Its Overexpression Confers Drought Tolerance in Arabidopsis. Plant Cell Environ. 2014, 37, 1097–1113.
- Yoo, Y.H.; Nalini Chandran, A.K.; Park, J.C.; Gho, Y.S.; Lee, S.W.; An, G.; Jung, K.H. OsPhyB-Mediating Novel Regulatory Pathway for Drought Tolerance in Rice Root Identified by a Global RNA-Seq Transcriptome Analysis of Rice Genes in Response to Water Deficiencies. Front. Plant Sci. 2017, 8, 580.
- Abbaraju, H.K.R.; Gupta, R.; Appenzeller, L.M.; Fallis, L.P.; Hazebroek, J.; Zhu, G.; Bourett, T.M.; Howard, R.J.; Weers, B.; Lafitte, R.H.; et al. A vegetative storage protein improves drought tolerance in maize. Plant Biotechnol. J. 2021.
- Wen, Z.; Wang, Y.; Xia, C.; Zhang, Y.; Zhang, H. Chloroplastic SaNADP-ME4 of C(3)-C(4) Woody Desert Species Salsola laricifolia Confers Drought and Salt Stress Resistance to Arabidopsis. Plants 2021, 10, 1827.
- Gómez-Ocampo, G.; Ploschuk, E.L.; Mantese, A.; Crocco, C.D.; Botto, J.F. BBX21 Reduces Abscisic Acid Sensitivity, Mesophyll Conductance and Chloroplast Electron Transport Capacity to Increase Photosynthesis and Water Use Efficiency in Potato Plants Cultivated Under Moderated Drought. Plant J. 2021, 108, 1131–1144.
- Akimoto-Tomiyama, C.; Tanabe, S.; Kajiwara, H.; Minami, E.; Ochiai, H. Loss of Chloroplast-Localized Protein Phosphatase 2Cs in Arabidopsis thaliana Leads to Enhancement of Plant Immunity and Resistance to Xanthomonas campestris pv. campestris Infection. Mol. Plant Pathol. 2018, 19, 1184–1195.
- Erickson, J.L.; Adlung, N.; Lampe, C.; Bonas, U.; Schattat, M.H. The Xanthomonas effector XopL Uncovers the Role of Microtubules in Stromule Extension and Dynamics in Nicotiana benthamiana. Plant J. 2018, 93, 856–870.
- Gou, J.Y.; Li, K.; Wu, K.; Wang, X.; Lin, H.; Cantu, D.; Uauy, C.; Dobon-Alonso, A.; Midorikawa, T.; Inoue, K.; et al. Wheat Stripe Rust Resistance Protein WKS1 Reduces the Ability of the Thylakoid-Associated Ascorbate Peroxidase to Detoxify Reactive Oxygen Species. Plant Cell 2015, 27, 1755–1770.
- Wang, S.; Li, Q.P.; Wang, J.; Yan, Y.; Zhang, G.L.; Yan, Y.; Zhang, H.; Wu, J.; Chen, F.; Wang, X.; et al. YR36/WKS1-Mediated Phosphorylation of PsbO, an Extrinsic Member of Photosystem II, Inhibits Photosynthesis and Confers Stripe Rust Resistance in Wheat. Mol. Plant 2019, 12, 1639–1650.
- Hamel, L.P.; Sekine, K.T.; Wallon, T.; Sugiwaka, Y.; Kobayashi, K.; Moffett, P. The Chloroplastic Protein THF1 Interacts with the Coiled-Coil Domain of the Disease Resistance Protein N’ and Regulates Light-Dependent Cell Death. Plant Physiol. 2016, 171, 658–674.
- Jiang, G.; Yin, D.; Zhao, J.; Chen, H.; Guo, L.; Zhu, L.; Zhai, W. The Rice Thylakoid Membrane-Bound Ascorbate Peroxidase OsAPX8 Functions in Tolerance to Bacterial Blight. Sci. Rep. 2016, 6, 26104.
- Liu, M.; Zhang, S.; Hu, J.; Sun, W.; Padilla, J.; He, Y.; Li, Y.; Yin, Z.; Liu, X.; Wang, W.; et al. Phosphorylation-Guarded Light-Harvesting complex II Contributes to Broad-Spectrum Blast Resistance in Rice. Proc. Natl. Acad. Sci. USA 2019, 116, 17572–17577.
- Ishiga, Y.; Ishiga, T.; Ikeda, Y.; Matsuura, T.; Mysore, K.S. NADPH-Dependent Thioredoxin Reductase C Plays a Role in Nonhost Disease Resistance Against Pseudomonas syringae Pathogens by Regulating Chloroplast-Generated Reactive Oxygen Species. Peer. J. 2016, 4, e1938.
- Su, J.; Yang, L.; Zhu, Q.; Wu, H.; He, Y.; Liu, Y.; Xu, J.; Jiang, D.; Zhang, S. Active Photosynthetic Inhibition Mediated by MPK3/MPK6 is Critical to Effector-Triggered Immunity. PLoS Biol. 2018, 16, e2004122.
- Rodríguez-Herva, J.J.; González-Melendi, P.; Cuartas-Lanza, R.; Antúnez-Lamas, M.; Río-Alvarez, I.; Li, Z.; López-Torrejón, G.; Díaz, I.; Del Pozo, J.C.; Chakravarthy, S.; et al. A Bacterial Cysteine Protease Effector Protein Interferes with Photosynthesis to Suppress Plant Innate Immune Responses. Cell Microbiol. 2012, 14, 669–681.
- Wangdi, T.; Uppalapati, S.R.; Nagaraj, S.; Ryu, C.M.; Bender, C.L.; Mysore, K.S. A Virus-Induced Gene Silencing Screen Identifies a Role for Thylakoid Formation1 in Pseudomonas syringae pv Tomato Symptom Development in Tomato and Arabidopsis. Plant Physiol. 2010, 152, 281–292.
- Nakano, M.; Mukaihara, T. Ralstonia solanacearum Type III Effector RipAL Targets Chloroplasts and Induces Jasmonic Acid Production to Suppress Salicylic Acid-Mediated Defense Responses in Plants. Plant Cell Physiol. 2018, 59, 2576–2589.
- Faris, J.D.; Zhang, Z.; Lu, H.; Lu, S.; Reddy, L.; Cloutier, S.; Fellers, J.P.; Meinhardt, S.W.; Rasmussen, J.B.; Xu, S.S.; et al. A Unique Wheat Disease Resistance-like Gene Governs Effector-triggered Susceptibility to Necrotrophic Pathogens. Proc. Natl. Acad. Sci. USA 2010, 107, 13544–13549.
- Gao, C.; Xu, H.; Huang, J.; Sun, B.; Zhang, F.; Savage, Z.; Duggan, C.; Yan, T.; Wu, C.H.; Wang, Y.; et al. Pathogen Manipulation of Chloroplast Function Triggers a Light-dependent Immune Recognition. Proc. Natl. Acad. Sci. USA 2020, 117, 9613–9620.
- Tang, L.; Yang, G.; Ma, M.; Liu, X.; Li, B.; Xie, J.; Fu, Y.; Chen, T.; Yu, Y.; Chen, W.; et al. An Effector of a Mecrotrophic Fungal Pathogen Targets the Calcium-sensing Receptor in Chloroplasts to Inhibit Host Resistance. Mol. Plant Pathol. 2020, 21, 686–701.
- Wang, Q.L.; Chen, J.H.; He, N.Y.; Guo, F.Q. Metabolic Reprogramming in Chloroplasts under Heat Stress in Plants. Int. J. Mol. Sci. 2018, 19, 849.
- Yoo, Y.H.; Hong, W.J.; Jung, K.H. A Systematic View Exploring the Role of Chloroplasts in Plant Abiotic Stress Responses. Biomed. Res Int. 2019, 2019, 6534745.
- Rossi, S.; Burgess, P.; Jespersen, D.; Huang, B. Heat-Induced Leaf Senescence Associated with Chlorophyll Metabolism in Bentgrass Lines Differing in Heat Tolerance. Crop Sci. 2017, 57, S169–S178.
- Jespersen, D.; Zhang, J.; Huang, B. Chlorophyll Loss Associated with Heat-induced Senescence in Bentgrass. Plant Sci. 2016, 249, 1–12.
- Li, Z.; Tang, M.; Hassan, M.J.; Zhang, Y.; Han, L.; Peng, Y. Adaptability to High Temperature and Stay-Green Genotypes Associated with Variations in Antioxidant, Chlorophyll Metabolism, and γ-Aminobutyric Acid Accumulation in Creeping Bentgrass Species. Front. Plant Sci. 2021, 12, 750728.
- Jiao, B.; Meng, Q.; Lv, W. Roles of Stay-green (SGR) Homologs during Chlorophyll Degradation in Green Plants. Bot. Stud. 2020, 61, 25.
- Ono, K.; Kimura, M.; Matsuura, H.; Tanaka, A.; Ito, H. Jasmonate Production through Chlorophyll A Degradation by Stay-Green in Arabidopsis thaliana. J. Plant Physiol. 2019, 238, 53–62.
- Shimoda, Y.; Ito, H.; Tanaka, A. Arabidopsis STAY-GREEN, Mendel’s Green Cotyledon Gene, Encodes Magnesium-Dechelatase. Plant Cell 2016, 28, 2147–2160.
- Danilova, M.N.; Kudryakova, N.V.; Andreeva, A.A.; Doroshenko, A.S.; Pojidaeva, E.S.; Kusnetsov, V.V. Differential Impact of Heat Stress on the Expression of Chloroplast-Encoded Genes. Plant Physiol. Biochem. 2018, 129, 90–100.
- Chovancek, E.; Zivcak, M.; Botyanszka, L.; Hauptvogel, P.; Yang, X.; Misheva, S.; Hussain, S.; Brestic, M. Transient Heat Waves May Affect the Photosynthetic Capacity of Susceptible Wheat Genotypes Due to Insufficient Photosystem I Photoprotection. Plants 2019, 8, 282.
- Vacca, R.A.; Valenti, D.; Bobba, A.; Merafina, R.S.; Passarella, S.; Marra, E. Cytochrome C is Released in a Reactive Oxygen Species-Dependent Manner and is Degraded Via Caspase-like Proteases in Tobacco Bright-Yellow 2 Cells En Route to Heat Shock-Induced Cell Death. Plant Physiol. 2006, 141, 208–219.
- Martins, M.Q.; Rodrigues, W.P.; Fortunato, A.S.; Leitão, A.E.; Rodrigues, A.P.; Pais, I.P.; Martins, L.D.; Silva, M.J.; Reboredo, F.H.; Partelli, F.L.; et al. Protective Response Mechanisms to Heat Stress in Interaction with High Conditions in Coffea spp. Front Plant Sci. 2016, 7, 947.
- Cen, W.; Liu, J.; Lu, S.; Jia, P.; Yu, K.; Han, Y.; Li, R.; Luo, J. Comparative Proteomic Analysis of QTL CTS-12 Derived from Wild Rice (Oryza rufipogon Griff.), in the Regulation of Cold Acclimation and De-Acclimation of Rice (Oryza sativa L.) in Response to Severe Chilling Stress. BMC Plant Biol. 2018, 18, 163.
- Hong, W.J.; Jiang, X.; Ahn, H.R.; Choi, J.; Kim, S.R.; Jung, K.H. Systematic Analysis of Cold Stress Response and Diurnal Rhythm Using Transcriptome Data in Rice Reveals the Molecular Networks Related to Various Biological Processes. Int. J. Mol. Sci. 2020, 21, 6872.
- Liu, X.; Zhou, Y.; Xiao, J.; Bao, F. Effects of Chilling on the Structure, Function and Development of Chloroplasts. Front. Plant Sci. 2018, 9, 1715.
- Skupień, J.; Wójtowicz, J.; Kowalewska, Ł.; Mazur, R.; Garstka, M.; Gieczewska, K.; Mostowska, A. Dark-Chilling Induces Substantial Structural Changes and Modifies Galactolipid and Carotenoid Composition During Chloroplast Biogenesis in Cucumber (Cucumis sativus L.) Cotyledons. Plant Physiol. Biochem. 2017, 111, 107–118.
- Xue, M.; Guo, T.; Ren, M.; Wang, Z.; Tang, K.; Zhang, W.; Wang, M. Constitutive Expression of Chloroplast Glycerol-3-Phosphate Acyltransferase from Ammopiptanthus Mongolicus Enhances Unsaturation of Chloroplast Lipids and Tolerance to Chilling, Freezing and Oxidative Stress in Transgenic Arabidopsis. Plant Physiol. Biochem. 2019, 143, 375–387.
- Shi, H.; Ye, T.; Zhong, B.; Liu, X.; Chan, Z. Comparative Proteomic and Metabolomic Analyses Reveal Mechanisms of Improved Cold Stress Tolerance in Bermudagrass (Cynodon dactylon (L.) Pers.) by Exogenous Calcium. J. Integr. Plant Biol. 2014, 56, 1064–1079.
- van Buer, J.; Cvetkovic, J.; Baier, M. Cold Regulation of Plastid Ascorbate Peroxidases Serves as A Priming Hub Controlling ROS Signaling in Arabidopsis thaliana. BMC Plant Biol. 2016, 16, 163.
- Zhao, H.; Ye, L.; Wang, Y.; Zhou, X.; Yang, J.; Wang, J.; Cao, K.; Zou, Z. Melatonin Increases the Chilling Tolerance of Chloroplast in Cucumber Seedlings by Regulating Photosynthetic Electron Flux and the Ascorbate-Glutathione Cycle. Front. Plant Sci. 2016, 7, 1814.
- Soliman, M.H.; Alayafi, A.A.M.; El Kelish, A.A.; Abu-Elsaoud, A.M. Acetylsalicylic Acid Enhance Tolerance of Phaseolus vulgaris L. to Chilling Stress, Improving Photosynthesis, Antioxidants and Expression of Cold Stress Responsive Genes. Bot. Stud. 2018, 59, 6.
More
