Human coronaviruses (HCoVs) are associated with a range of respiratory symptoms. The discovery of severe acute respiratory syndrome (SARS)-CoV, Middle East respiratory syndrome, and SARS-CoV-2 pose a significant threat to human health. The HCoV-MS method is a sensitive assay that combines multiplex PCR with matrix-assisted laser desorption/ionization-time of flight mass spectrometry (MALDI-TOF MS), to detect and differentiate seven HCoVs simultaneously.
- human coronavirus
- MALDI-TOF MS
- RT-PCR
- high throughput
1. Introduction
Coronaviruses (CoVs) are large, enveloped, positive-sense RNA viruses that cause respiratory diseases in a range of animals, including humans [1]. CoVs are divided into four genera, namely δ-CoVs, γ-CoVs, β-CoVs, and α-CoVs, among which β-CoVs and α-CoVs can infect mammals [2]. Seven human CoVs (HCoVs) have been identified, including HCoV-NL63, HCoV-229E, HCoV-OC43, HCoV-HKU1, SARS-CoV, MERS-CoV, and SARS-CoV-2 [1][3][1,3]. Respiratory diseases caused by HCoV infection range from mild to severe. Approximately 15–30% of respiratory tract infections worldwide each year are caused by HCoV-229E, HCoV-OC43, HCoV-NL63, and HCoV-HKU1. They are mild and self-healing diseases that do not pose a major threat to public health [4].
2. Performance of the Hcov-MS Method
3. Specificity of the HCoV-MS Method
4. Sensitivity of the HCoV-MS Method
Serial plasmid dilutions were used to evaluate the sensitivity of the HCoV-MS method. The detection limits of part of the target genes are shown in Figure 12, and listed in Table 12. The detection limit of the HCoV-MS method was found to be 1–5 copies/reaction.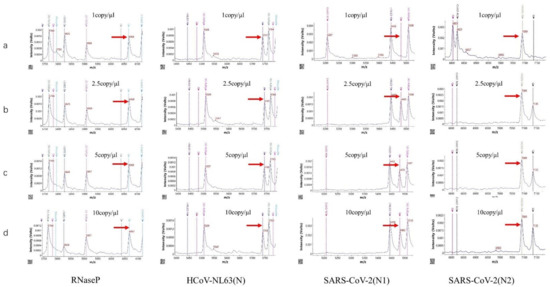
| Assays | Target | Detection Limit (Copies/Reaction) |
|---|---|---|
| 5 | ||
| SARS-CoV-2 | ||
| N1 | ||
| 2.5 | ||
| N2 | ||
| 2.5 | ||
| S | 2.5 | |
| ORF1b | 2.5 |
5. Sensitivity Comparisons of HCoV-MS and RT-PCR
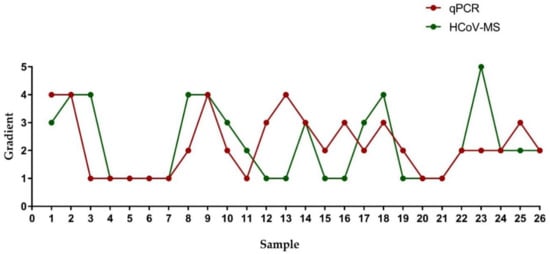
| Sample | Gradient | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | |||||||||
| RT-PCR * | HCoV-MS * | RT-PCR | HCoV-MS | RT-PCR | HCoV-MS | RT-PCR | HCoV-MS | RT-PCR | HCoV-MS | ||||
| Human RNase P | Human RNase P | 1 | |||||||||||
| 1 | No Ct/No Ct | − 1 | No Ct/No Ct | − | 40.74/No Ct | + 2 | 36.87/No Ct | + | 34.17/No Ct | HCoV-NL63 | N | 1 | |
| + | |||||||||||||
| 2 | No Ct/No Ct | − | No Ct/No Ct | − | 40.65/No Ct | − | 36.30/No Ct | + | 34.36/No Ct | RdRp | 1 | ||
| + | HCoV-229E | N | 2.5 | ||||||||||
| 3 | 35.94/No Ct | − | 32.84/No Ct | − | 33.35/No Ct | − | 32.56/No Ct | + | 30.16/No Ct | + | |||
| 4 | 36.88/34.01 | + | 35.96/32.03 | + | 32.92/29.77 | + | 31.24/27.63 | + | 28.52/25.92 | + | RdRp | 2.5 | |
| 5 | 39.64/33.68 | + | 37.01/32.01 | + | 34.79/30.05 | + | 33.03/27.92 | + | 30.6/25.83 | + | HCoV-OC43 | N | 2.5 |
| 6 | 37.33/35.04 | + | 36.67/32.81 | + | 35.45/31.17 | + | 34.28/29.21 | RdRp | 2.5 | ||||
| + | 32.9/27.14 | + | HCoV-HKU1 | N | |||||||||
| 7 | 40.63/34.79 | + | 40.11/32.60 | + | 38.66/30.97 | + | 35.83/28.46 | + | 33.83/26.86 | + | 1 | ||
| 8 | No Ct/No Ct | − | No Ct/39.02 | − | 39.35/36.45 | − | 36.3/34.71 | + | 35.15/32.30 | + | RdRp | 2.5 | |
| 9 | No Ct/No Ct | − | No Ct/No Ct | − | No Ct/No Ct | − | 39.33/36.50 | + | 36.32/34.60 | + | MERS-CoV | N | 1 |
| 10 | No Ct/No Ct | − | 37.23/37.43 | − | 37.06/34.76 | + | 35.16/32.90 | + | 33.24/30.99 | + | RdRp | 2.5 | |
| 11 | 39.02/36.78 | − | 36.35/33.84 | + | 34.35/31.44 | + | 32.36/30.16 | + | 30.66/28.16 | + | E | 2.5 | |
| 12 | No Ct/No Ct | + | No Ct/No Ct | + | No Ct/38.39 | + | 35.95/35.15 | + | 34.61/32.06 | ORF1b | 2.5 | ||
| SARS-CoV | E | 5 | |||||||||||
| + | |||||||||||||
| 13 | No Ct/No Ct | + | No Ct/No Ct | + | No Ct/No Ct | + | 34.99/No Ct | + | 33.94/38.01 | + | |||
| 14 | No Ct/No Ct | − | No Ct/No Ct | − | No Ct/38.16 | + | 37.12/35.66 | + | 34.52/33.22 | + | N | 5 | |
| 15 | No Ct/No Ct | + | 37.15/36.10 | + | 37.04/33.67 | + | 35.26/31.76 | + | 33.01/30.57 | + | ORF1b | ||
| 16 | No Ct/No Ct | + | 40.81/No Ct | + | No Ct/38.45 | + | 39.89/36.62 | + | 37.84/35.04 | + | |||
| 17 | No Ct/No Ct | − | No Ct/39.73 | − | 39.31/37.82 | + | 37.87/36.36 | + | 36.43/34.64 | + | |||
| 18 | No Ct/No Ct | − | No Ct/40.08 | − | No Ct/39.48 | − | 42.18/38.98 | + | 40.41/36.18 | + | |||
| 19 | No Ct/No Ct | + | 37.22/36.00 | + | 36.90/34.46 | + | 34.81/33.69 | + | 32.92/31.26 | + | |||
| 20 | 37.80/32.63 | + | 36.22/30.56 | + | 33.91/28.28 | + | 32.24/26.46 | + | 29.53/23.98 | + | |||
| 21 | 39.93/34.69 | + | 36.10/32.38 | + | 32.28/30.01 | + | 30.51/27.60 | + | 27.69/25.37 | + | |||
| 22 | No Ct/No Ct | − | No Ct/38.56 | + | 38.56/39.32 | + | 37.10/34.75 | + | 36.13/33.54 | + | |||
| 23 | No Ct/No Ct | − | 35.15/No Ct | − | 34.93/No Ct | − | 34.15/38.72 | − | 30.79/35.09 | + | |||
| 24 | No Ct/No Ct | − | 39.17/36.95 | + | 36.61/34.35 | + | 35.95/31.70 | + | 33.32/29.73 | + | |||
| 25 | No Ct/No Ct | − | No Ct/No Ct | + | No Ct/38.88 | + | 36.66/37.20 | + | 33.94/35.70 | + | |||
| 26 | No Ct/No Ct | − | 36.46/No Ct | + | 34.97/38.94 | + | 35.39/35.67 | + | 33.65/34.16 | + | |||
6. Research Findings of the HCoV-MS Method
A HCoV-MS method was established to simultaneously detect seven HcoVs, which can be used as a detection system when new HCoVs appear. Notably, the detection sensitivity of HCoV-MS is 1–5 copies/reaction. Except for SARS-CoV, other HCoVs could be detected with 1–2.5 copies/reaction. HCoV-NL63 was even detected with 1 copy/reaction. This sensitivity of this method outperforms that of other detection methods [20][21][22][23][24][21,22,23,24,25].
When testing 151 unknown clinical samples, the specificity and sensitivity of the HCoV-MS method reached 100%, surpassing other methods [25][26][27][26,27,28]. This could, however, be due to insufficient clinical samples. Samples of human/animal throat swabs or cell cultures were obtained in cooperation with UN-CoV-2020. Seven samples were positive, including four SARS-CoV-2 samples, two SARS-CoV samples, and one HCoV-NL63 sample. The results obtained with the HCoV-MS method were consistent with the official answers (Table S3). Moreover, the concentration of individual positive samples was low, but could still be accurately identified, which further highlights the detection ability of the HCoV-MS method.
The HCoV-MS method is high throughput, as reflected in the detection of multiple target genes and the requirement of small sample sizes. The HCoV-MS method could simultaneously detect 384 targets in one run spanning 30 min, with results automatically determined by the relevant software. Moreover, the reagent cost of the HCoV-MS method is relatively low, making this method ideal for large-scale population screening.
The HCoV-MS also has some limitations. This method is difficult to identify new HCoV because it is based on comparing and analyzing known HCoV sequences, selecting gene fragments with conserved intraspecies specificity. In conclusion, the HCoV-MS method has the characteristics of high throughput, speed, and sensitivity, only requiring a small number of samples. Therefore, it is expected to be a supplement to real-time PCR technology.
