Your browser does not fully support modern features. Please upgrade for a smoother experience.
Please note this is a comparison between Version 1 by Chien-Hsiu Li and Version 2 by Catherine Yang.
The microRNA (miRNA) Let-7 has been identified as related to glycolysis procedures such as tissue repair, stem cell-derived cardiomyocytes, and tumoral metastasis. In many cancers, the expression of glycolysis-related enzymes is correlated with Let-7, in which multiple enzymes are related to the regulation of the autophagy process. However, much recent research has not comprehensively investigated how Let-7 participates in glycolytic reprogramming or its links to autophagic regulations, mainly in tumor progression.
- Let-7
- microRNA
- autophagy
1. Introduction
Cellular energy-related metabolisms involve complex regulation dynamic processes. The current understanding is that the uptake of glucose from the extracellular environment is a primary way for cells to acquire resources for sustaining energy. Intermediate glucose metabolism can be converted by diverse metabolites of lipids and amino acids to maintain cellular functions [1]. In addition, autophagy is recognized as a digesting process to engulf cellular compartments or damaged organelles for maintaining metabolic homeostasis while responding to multiple metabolic stresses [2]. Within such processes, necessary molecules can be recycled by degrading specific factors to adapt cell growth to a rigorous environment. The glucose metabolic networks regulated by glycolysis and autophagy have explained the fundamental nutrients dynamic for maintaining cell growth and survival. Among them, miRNA, a 18–25-nt single-stranded noncoding RNA, serves as an essential modulator involved in cellular metabolisms, conducting post-transcriptional modification by targeting to 3’UTR of specific mRNA [3].
Let-7 is the first miRNA family identified as involved in multiple cellular and biological functions, including glucose metabolism and autophagy. The glucose metabolism is controlled by the miRNA family of Let-7 directly [4], or regulated by an autophagy-associated glycogen recycling system [5][6][5,6]. The imbalance of Let-7-mediated processes of glucose metabolism has been found to contribute to disease progression, especially carcinogenesis. In addition, metabolic dysregulation, which causes excessive energy release for unlimited growth, has been a consequential risk for promoting cancer development. However, the crosstalk networks between autophagy and glucose metabolism—especially the linkage of Let-7 miRNA that participates in carcinogenesis and various biological functions—are still obscure and need to be fully addressed.
2. Involvement of Let-7 in Glycolysis Reprogramming
Let-7 was reported in 1990 and contributes to the embryonic development of C. elegans. The artificial manipulation of the expression of Let-7 causes mortality during embryogenesis [7]. Interestingly, several cancer-associated molecules have been identified from embryonic development, including Let-7. The Let-7 family has been classified by its consensus sequence [8] (Table 1). According to the literatures review, the Let-7 family-related expression was associated with the patient’s prognosis (Table 2). Furthermore, numerous studies have indicated that the related expression of Let-7 is lower in tumor cells, whereas an increased level of Let-7 is able to suppress tumor malignancy, which indicates that Let-7 may contribute to the suppression role in most types of tumors [9][10][9,10].Table 1. The Let-7 family in humans.
| Let-7 Family | Sequence |
|---|---|
| Let-7a | UGAGGUAGUAGGUUGUAUAGUU |
| miR-202 | |
| AGAGGUAGUAGGGCAUGGGAA | |
Table 2. Let-7 family in pan-cancer to on the basis of literature review to coordinate the related survival correlation between patients with cancer and the Let-7 family.
| Cancer Type | Let-7 Family | Clinical Association | Year | Reference | |||
|---|---|---|---|---|---|---|---|
| Acute Myeloid Leukemia | Let-7a | Associated with poor outcome | 2013 | [11] | [16] | ||
| Let-7b | UGAGGUAGUAGGUUGUGUGGUU | ||||||
| Let-7a-2-3p | Associated with good outcome | 2015 | [12] | [17] | Let-7c | UGAGGUAGUAGGUUGUAUGGUU | |
| miR-98 | Associated with good outcome | 2019 | [13] | [18] | Let-7d | AGAGGUAGUAGGUUGCAUAGUU | |
| Breast Cancer | Let-7a | Associated with good outcome | 2018 | [14] | [19] | Let-7e | UGAGGUAGGAGGUUGUAUAGUU |
| Let-7a | Associated with good outcome | 2018 | [15] | [20] | Let-7f | UGAGGUAGUAGAUUGUAUAGUU | |
| Let-7a | Associated with good outcome | 2019 | [16] | [21] | Let-7g | UGAGGUAGUAGUUUGUACAGUU | |
| Let-7a | Associated with good outcome | 2019 | [17] | [22] | Let-7i | UGAGGUAGUAGUUUGUGCUGUU | |
| Let-7a-5p | Associated with good outcome | 2020 | [18] | [23] | miR-98 | ||
| Let-7b | UGAGGUAGUAAGUUGUAUUGUU | ||||||
| Associated with good outcome | 2018 | [ | 14] | [19] | |||
| Let-7b | Associated with good outcome | 2019 | [16] | [21] | |||
| Let-7b | Associated with good outcome | 2020 | [19] | [24] | |||
| Let-7b | Associated with good outcome | 2020 | [20] | [25] | |||
| Let-7b | Associated with good outcome | 2020 | [21] | [26] | |||
| Let-7b | Associated with good outcome | 2016 | [22] | [27] | |||
| Let-7c | Associated with good outcome | 2016 | [22] | [27] | |||
| Let-7c | Associated with good outcome | 2018 | [14] | [19] | |||
| Let-7c | Associated with good outcome | 2019 | [16] | [21] | |||
| Let-7c | Associated with poor outcome | 2020 | [23] | [28] | |||
| Let-7d | Associated with good outcome | 2018 | [14] | [19] | |||
| Let-7d | Associated with good outcome | 2018 | [24] | [29] | |||
| Let-7d | Associated with good outcome | 2019 | [16] | [21] | |||
| Let-7e | Associated with good outcome | 2018 | [14] | [19] | |||
| Let-7e | Associated with poor outcome | 2019 | [16] | [21] | |||
| Let-7f | Associated with good outcome | 2018 | [14] | [19] | |||
| Let-7f | Associated with good outcome | 2019 | [16] | [21] | |||
| Let-7g | Associated with good outcome | 2011 | [25] | [30] | |||
| Let-7g | Associated with good outcome | 2018 | [14] | [19] | |||
| Let-7g | Associated with good outcome | 2019 | [16] | [21] | |||
| Let-7i | Associated with good outcome | 2008 | [26] | [31] | |||
| Let-7i | Associated with good outcome | 2018 | [14] | [19] | |||
| Let-7i | Associated with good outcome | 2019 | [16] | [21] | |||
| Colon Cancer | Let-7a | Associated with poor outcome | 2017 | [27] | [32] | ||
| Let-7g | Associated with good outcome | 2017 | [28] | [33] | |||
| Esophageal Cancer | Let-7b | Associated with good outcome | 2012 | [29] | [34] | ||
| Let-7c | Associated with good outcome | 2012 | [29] | [34] | |||
| Let-7c | Associated with good outcome | 2013 | [30] | [35] | |||
| Glioblastoma | Let-7a | Associated with good outcome | 2013 | [31] | [36] | ||
| Let-7c | Associated with good outcome | 2021 | [32] | [37] | |||
| Let-7f | Associated with poor outcome | 2018 | [33] | [38] | |||
| Let-7i | Associated with good outcome | 2020 | [34] | [39] | |||
| Liver Cancer | Let-7a | Associated with poor outcome | 2018 | [35] | [40] | ||
| Let-7a | Associated with good outcome | 2020 | [36] | [41] | |||
| Let-7b | Associated with good outcome | 2020 | [36] | [41] | |||
| Let-7b | Associated with good outcome | 2020 | [37] | [42] | |||
| Let-7c | Associated with good outcome | 2020 | [36] | [41] | |||
| miR-202 | Associated with good outcome | 2020 | [38] | [43] | |||
| Lung Adenocarcinoma | Let-7b | Associated with good outcome | 2021 | [39] | [44] | ||
| Melanoma | miR-98 | Associated with good outcome | 2014 | [40] | [45] | ||
| Mesothelioma | Let-7c | Associated with good outcome | 2017 | [41] | [46] | ||
| Ovarian Cancer | Let-7b | Associated with poor outcome | 2021 | [42] | [47] | ||
| Let-7d | Associated with poor outcome | 2012 | [43] | [48] | |||
| Let-7e | Associated with good outcome | 2017 | [44] | [49] | |||
| Let-7f | Associated with good outcome | 2013 | [45] | [50] | |||
| Let-7g | Associated with poor outcome | 2016 | [46] | [51] | |||
| Let-7i | Associated with good outcome | 2008 | [26] | [31] | |||
| miR-98 | Associated with good outcome | 2021 | [47] | [52] | |||
| miR-98 | Associated with good outcome | 2020 | [48] | [53] | |||
| miR-98 | Associated with poor outcome | 2019 | [49] | [54] | |||
| miR-98 | Associated with poor outcome | 2018 | [50] | [55] | |||
| miR-202 | Associated with good outcome | 2020 | [51] | [56] | |||
| Let-7g | Associated with good outcome | 2017 | [52] | [57] | |||
| Pancreatic Cancer | Let-7e | Associated with good outcome | 2010 | [53] | [58] | ||
| miR-202 | Associated with good outcome | 2021 | [54] | [59] | |||
| Prostate Cancer | Let-7b | Associated with poor outcome | 2013 | [55] | [60] | ||
| Let-7c | Associated with good outcome | 2013 | [55] | [60] |
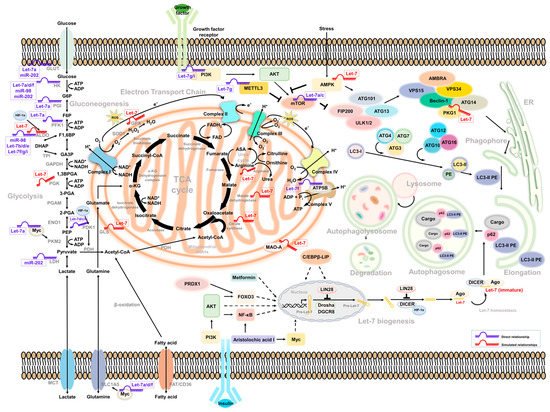
Figure 1.
Intermediate mediators/molecules between Let-7-associated glucose metabolism and autophagy.
3. Let-7-Mediated Autophagy Participates in Glucose Metabolism and Cancer Progression
3.1. Let-7 and Autophagy
In lung cancer, Let-7 targets IGF-1R to induce autophagy and blocks the function of BCL2L1/BCL2/PI3K complex to induce apoptosis and pyroptosis and inhibit cell motility [61][88]. Let-7a targets Rictor’s mTORC2 component, inhibiting AKT/mTORC1 signaling to activate autophagy in gastric cancer [62][89]. Similar regulation can be observed in human placental trophoblasts, in which the expression of Let-7b was correlated with cell growth and motility. The Let-7b-mediated TGFBR1/ERK/IL-6/TNF-α cascade triggers not only apoptosis but also autophagy. Such regulation may contribute to pre-eclampsia during pregnancy [63][90]. In glioma, the downregulation of STAT3 was mediated by Let-7a, Let-7d, and Let-7f. Upregulation of Let-7 suppressed the expression of STAT3, resulting in the inhibition of cell proliferation and induction of autophagy and apoptosis [64][91]. Liang et al. identified that a set of the Let-7 family was downregulated in hepatocellular carcinoma, with different clinical correlations under a genetic profiling analysis. The expression of Let-7b and Let-7c had a better prognosis; Let-7e had a poor prognosis instead. Among them, Let-7e has been demonstrated to promote tumor growth by suppressing autophagy and apoptosis [65][92]. A similar strategy was used in cholangiocarcinoma. Clinical evidence showed that the expression of NUAK1 was negatively correlated with Let-7a. NUAK1-mediated cholangiocarcinoma cell motility can be suppressed by increasing Let-7a. In turn, the overexpression of Let-7a inhibited NUAK1-mediated tumor malignancy by the induction of autophagy [66][93]. Additionally, Let-7 can be regulated by LncRNA H19 and LIN28 in breast cancer. The expression of long non-coding RNA (lncRNA) H19 and LIN28 was correlated with breast cancer’s poor prognosis and metastasis ability. Overexpression of H19 and LIN28 increases the expression of several autophagy-related ATG markers as well as its puncta structure formation. Downregulation of Let-7 increased the transcript activity of several EMT-related genes—including Slug, Zeb1, Twist, Snail, β-catenin, and HMGA2—to modulate the metastasis of breast cancer [67][94]. Another lncRNA MIR99AHG, as well as its Let-7c-associated cluster, were reported to have decreased expression in lung cancer. MIR99AHG increased Let-7c, subsequently promoting autophagy via targeting mTOR, an autophagy suppressor of nucleation, and ANXA2, a negative regulator of elongation, to suppress the growth and motility of lung adenocarcinoma [68][95]. In view of the controversial role of autophagy in a variety of cancers, the regulation of Let-7-mediated autophagy in tumor progression could be complicated—and condition-, environment-, and tissue-specific.
3.2. Autophagy Activators
Several components have been identified as triggering Let-7-mediated autophagy in cancer cells. Treating cells with recombinant capsid protein viral particle 1 (rVP1) induces autophagy to regulate the motility of macrophages [69][96] and ovarian cancer cells [70][97]. In ovarian cancer, autophagy—activated by either a canonical or a rVP1-mediated noncanonical pathway—maintains the homeostasis of the Let-7 level through SQSTM1-mediated degradation of Dicer/AGO2 inhibition of cell migration [70][97]. In lymphosarcoma, the expression of Let-7g/CTSB may be suppressed by ribonuclease binase to participate in apoptosis and autophagy [71][98].
3.3. Drug Resistance
In gastric cancer, the expression of miR-202 can be restricted by lncRNA MALAT1, resulting in the activation of autophagy, increased tumor malignancy, and an enhanced drug-resistant ability [72][99]. In agreement with other reports, Yang et al. showed that paclitaxel-based drug-resistant breast cancer cells express a high level of CircRNA ABCB10 and autophagy, which are correlated with clinical paclitaxel-sensitive or resistant data and negatively associated to Let-7a. Mechanistically, the Let-7a/DUSP7 axis is a downstream effector of Circ-ABCB10 resistant to paclitaxel treatment. Knockdown of Circ-ABCB10 not only increases sensitivity to paclitaxel but also decreases tumor weight [73][100]. Similar regulation was observed in a cisplatin-based resistance model of A549 with a high level of DICER. Overexpression of DICER induces autophagy processes and increased tumor growth and motility, in which DICER-mediated suppression of Let-7i and the PI3K/AKT/mTOR axis contributes to the autophagy activity [74][67]. In medulloblastoma, inhibited autophagy was found to promote tumor resistance upon cisplatin treatment. The level of Let-7f in cells was insufficient to repress HMGB1 and led to autophagy-mediated drug resistance. Overexpression of Let-7f could attenuate cisplatin’s drug resistance and induce apoptosis in medulloblastoma cells [75][101].
3.4. Let-7-Mediated Autophagy in Glucose Metabolism
Recently, Let-7-mediated autophagy has been described as participating in glucose metabolism events. For example, Duan et al. observed that Let-7 targeted BCL-xL to induce autophagic cell death in lung cancer, indicating that Let-7 regulates mitochondria-related autophagy (mitophagy) to regulate metabolism-related events, and BCL-xL with non-apoptotic functions to induce cell death [76][102]. However, the underlying mechanism of Let-7-mediated autophagy in glucose metabolism that contributes to cell stress and death needs to be further elucidated. According to the above reports, several links may support the correlation between Let-7, autophagy, and glucose metabolism. In turn, Lai et al. found that—in a hypoxic environment—HIF-1α can interact with DICER to regulate miRNA processing in diverse cancer types, including colon, breast, liver, lung, and prostate cancer [77][103]. HIF-1α changed the glycolysis-related enzyme PDK1 level and induced autophagy-mediated proteolysis by interacting with Parkin/p62 to possess DICER, which decreased Let-7 biogenesis. Overexpression of HIF-1α reduced the levels of Let-7a, Let-7b, and Let-7d as well as its complement downstream target LIN41 and Aurora B to promote tumor metastasis [77][103]. However, how glycolysis participates in DICER ubiquitination and related autophagy processes has not yet been well explained. So far, Lai et al.’s study provides a possible reason for why the Let-7 level being downregulated under hypoxia is an important factor contributing to tumor microenvironment reprogramming and providing tumor cells with escape from immune surveillance. Recently, bone marrow-derived human mesenchymal stem cells (hMSCs) have been observed to have anticancer activity. Egea et al. found that Let-7f can be transactivated under hypoxia to induce autophagy in hMSCs and promote migration in tumor cells [78][104]. Let-7f can be regulated by TGF-β, TNF-α, IL-1β, and SDF-1α to modulate CXCR4 and MMP-9 expression and drive chemotactic invasion. Interestingly, hMSCs have been observed to transport Let-7f by exosome secretion to inhibit the growth and motility of breast cancer; such events can be reversed with the Let-7f inhibitor [78][104].
3.5. mTOR-Dependent Autophagy and Glucose Metabolism
Several studies have found that Let-7 mediates glucose metabolism through the regulation of mammalian target of rapamycin (mTOR) [74][79][80][62][67,68,76,89]. It is also a well-known negative regulator of autophagy. Notably, human growth hormone receptors (GHR) have played an essential role in glucose metabolism and are linked to mTOR activity. A murine model revealed that, under limited nutrients, growth hormone maintained the cellular glucose level through gluconeogenesis, accompanied by the induction of autophagy [81][105]. In addition, GHR has been identified to contribute to breast and prostate cancer malignancy [82][83][106,107]. Elzein et al. reported that GHR is the target of miR-202. Increased miR-202 suppresses the expression of GHR in MCF and LNCaP cells [84][108]. Additionally, it has been reported that PKM2 and mTOR expression is downregulated under glucose restriction in breast cancer, which reverses the Warburg effect of cells [85][109]. Strikingly, these molecules were all be Let-7 downstream effectors. Such regulation may explain how Let-7 mediates autophagy and glucose metabolism to regulate cancer cell progression (Figure 1).
