Covalent crosslinks within or between proteins play a key role in determining the structure and function of proteins. Some of these are formed intentionally by either enzymatic or molecular reactions and are critical to normal physiological function. Others are generated as a consequence of exposure to oxidants (radicals, excited states or two-electron species) and other endogenous or external stimuli, or as a result of the actions of a number of enzymes (e.g., oxidases and peroxidases). Increasing evidence indicates that the accumulation of unwanted crosslinks, as is seen in ageing and multiple pathologies, has adverse effects on biological function.
1. Introduction
The formation of covalently linked peptides and proteins plays a key role in many biological processes, both physiologically and pathologically. These can be formed intentionally, such as in the oxidative folding of nascent proteins within mammalian cells in the endoplasmic reticulum or Golgi involving the generation of disulfide bonds from two cysteine (Cys) residues and in the assembly of insect exoskeletons via the crosslinking of two tyrosine (Tyr) residues, or as a result of accidental exposure to oxidizing species (low-molecular mass or enzymes) that chemically link two protein sites. These crosslinks can be formed between different sites within the same molecule (intramolecular or intrachain crosslinks), between two different chains in a single molecule (e.g., the interchain crosslinks in mammalian insulins), or between two separate species (intermolecular crosslinks). Some of these crosslinks play a key role in stabilizing or maintaining proteins structures and can be essential to functional activity
[1], whereas others have negative effects of biological function (e.g., altered turnover, lifetime or activity)
[2]. Whilst some crosslinks appear to be benign and devoid of adverse effects and end up as targets of catabolic processes (e.g., degradation by proteasomes, lysosomes, other proteases), others are strongly associated with adverse effects and are implicated (in some cases, causally) in the development of pathologies (e.g.,
[3][4][3,4]).
2. Enzymatic Protein Crosslinking
Multiple enzymes can mediate the crosslinking of proteins, with a few key examples briefly summarized below. Enzyme-generated crosslinks are critical to the formation of many three-dimensional structures as these provide strength and rigidity, if biologically required. Examples include crosslinks formed within the extracellular matrix (ECM) of most, if not all, tissues, such as those formed between matrix proteins, and particularly collagens by the copper-containing lysyl oxidase (LOX) and LOX-like (LOXL) enzymes [5](reviewed in [11]). LOX oxidizes specific lysine (Lys) and hydroxylysine residues to carbonyls that undergo subsequent reactions to crosslink collagens (e.g., types I and III) and elastin [5][6][7][8][11,12,13,14]. In contrast, the LOXL family of enzymes acts on collagen type IV and drives the assembly of basement membranes [5][9][11,15]. Other enzymes also contribute to collagen crosslinking in the ECM with peroxidasin, a member of the heme peroxidase superfamily, mediating the formation of highly specific methionine (Met) to Lys crosslinks within the NC1 domains on collagen via generation of the oxidant hypobromous acid (HOBr). This species reacts rapidly with the Met residue to form an intermediate that then reacts with a suitably positioned Lys residue [10][11][16,17] (see also below). This type of crosslinking has been reported across many species [12][18]. Other members of the peroxidase superfamilies (e.g., horseradish peroxidase, myeloperoxidase, laccase) can also generate crosslinks via enzyme-mediated oxidation of substrates to radicals which then undergo radical–radical coupling. A classic example is oxidative coupling of Tyr and a wide range of other phenols via phenoxyl radical generation [13][14][15][19,20,21].
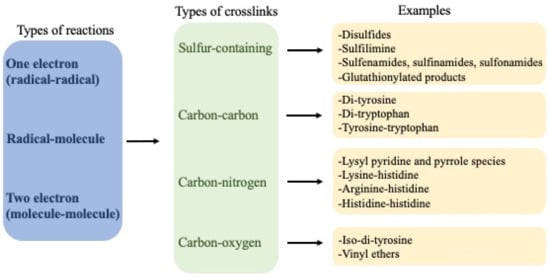
An overview of the crosslinks is presented in
Figure 1.
Figure 1.
Overview of crosslinks formed on proteins, their nature and mechanisms of formation.
3. One-Electron (Radical–Radical) Reactions
Dimerization of two radicals to form a new covalent bond is typically a very fast process due to the low energy barriers for such reactions. Therefore, they are a major source of crosslinks in peptides and proteins when the radical flux is high and there are limited competing reactions. Most carbon-centered protein radicals (P•) formed from aliphatic side-chains by hydrogen–atom abstraction reactions react rapidly with O2 at diffusion-controlled rates (k ~ 109 M−1 s−1) to give peptide or protein peroxyl radicals (P-OO•) [16][31]. The rapidity of these reactions limits direct reactions of two P•, except in circumstances where the O2 concentration is low. This is of biological relevance, as hypoxia is a common phenomenon, with endogenous levels of O2 being typically in the range 3–70 μM [17][32]. However, lower concentrations are present in situations where demand is great (e.g., high metabolic rates) or perfusion is poor (e.g., in the core of many solid tumors), thereby limiting P-OO• formation and allowing (P-P) dimer formation [18][33]. For the limited number of P•, where reaction with O2 is slow or modest, as is the case for Cys-derived thiyl radicals (RS•, k < 107 M−1 s−1 [19][34]), tryptophan (Trp) indolyl radicals (Trp•, k < 4 × 106 M−1 s−1 [20][21][35,36]) and Tyr phenoxyl radicals (Tyr•, k < 103 M−1 s−1 [22][37]), formation of disulfides (cystine) from two RS•, di-tyrosine from two Tyr•, di-tryptophan from two Trp•, and crossed dimers between these (e.g., Tyr–Trp) can be generated.
Light, particularly of wavelengths >~280 nm, which are not absorbed by the ozone layer, can penetrate significantly into biological structures and be absorbed either directly by protein residues, particularly Trp, Tyr and cystine [23][38], or by other species with high extinction coefficients in the long wavelength UV or visible regions. Energy absorption by non-protein species can give rise to indirect protein oxidation via the formation of excited states (e.g., singlet oxygen, 1O2 and reactive triplets) and/or radicals [23][38]. Direct UV absorption by proteins can form RS• from homolysis of the –S–S– bond of cystine (with C-S cleavage being an alternative pathway), and Tyr and Trp radicals by photo-ionization of these side-chains. These species can then give rise to crosslinks.
4. Radical–Molecule Reactions
Radical–molecule reactions appear to be a limited pathway for the formation of protein crosslinks, due to the absence of double bonds to which radicals might add in proteins, and limited stability of adducts to aromatic rings. Notable exceptions are the rare amino acids dehydroalanine (DHA; 2-aminoacrylic acid) and dehydroaminobutyric acid (DHB; 2-aminocrotonic acid). These contain a double bond between the α- and β-carbons of the side-chain and are non-proteinogenic species
[24][39], with these being generated via elimination reactions of serine residues (Ser), phospho-Ser and selenocysteine (Sec) residues (in the case of DHA)
[24][39], and from threonine (Thr) and phospho-Thr (in the case of DHB)
[25][40]. DHA can also be formed via cleavage of the carbon–sulfur bonds of the disulfide cystine, via mechanisms involving RS
• or nucleophilic elimination reactions
[24][39].
Although radical addition to double bonds is typically rapid and energetically favorable due to low energy barriers, these reactions are rare as the concentrations of both DHA and DHB (with the former more abundant) and the radicals that might undergo addition with them are very low. Nevertheless, some examples are known for radicals that have relatively long lifetimes and modest rates of reaction with O
2 (i.e., Cys thiyl, Tyr phenoxyl, Trp indolyl)
[26][41].
5. Two-Electron (Molecule–Molecule) Reactions
Reactions between two molecules are typically much slower than between two radicals or radical–molecule reactions. However, the concentration of the reactants is often much higher than for reactive intermediates, and consequently, the overall rates of these reactions may be significant—and the yield of products greater—than for the processes outlined above. These reactions are therefore major sources of protein crosslinks. The rate constants for these reactions would be expected to vary enormously—though quantitative data is lacking for most systems—with some reactions involving unstable species (e.g., sulfenic acids (RSOH), S-nitrosothiols (RSNO), unsaturated aldehydes/ketones, quinones) being relatively rapid (i.e., occurring over seconds/minutes).
6. Types of Crosslinks Detected within and between Proteins and Peptides
The following sections and
Table 1 summarize various types of crosslinks that have been detected within and between peptides and proteins, the nature of these species, their reversibility, mechanisms of formation and, subsequently, methods available to detect, identify, characterize and quantify these species.
Table 1. Examples of major non-disulfide protein crosslinks generated during non-enzymatic oxidative processes and methodologies employed to characterize them.
| Crosslinked Residues |
Protein(s) |
Chemical Nature and/or Mechanism of Formation of the Crosslink |
Method(s) |
Refs |
| Tyr-Cys |
|
- a)
-
Myoglobin
- b)
-
Galactose oxidase
- c)
-
Cysteine dioxygenase
|
|
|
- 1)
-
Michael addition from thiols (Cys) to oxidized Tyr species (a)
- 2)
-
Thioether bridge (C-S) (b and c)
|
|
Mass spectrometry (a)
X-ray crystallography (b, c) |
[27][28][29] | [42,43,44] |
| Trp-Cys |
Human growth hormone (hGH) |
|
- 1)
-
Michael addition from N (Trp indole) to DHA (formed from Cys)
- 2)
-
Thioether bridge (C-S)
|
|
Mass spectrometry |
[26] | [41] |
| Met-Hydroxy-lysine |
Collagen IV |
Formation of S=N bridge (sulfilimine bond) induced by peroxidasin/HOBr |
Mass spectrometry |
[11] | [17] |
| Lys-Cys |
Transaldolase |
Nitrogen–oxygen–sulfur (NOS) link/redox switch |
X-ray crystallography |
[30] | [45] |
| Cys-Ser |
|
- a)
-
Human growth hormone
- b)
-
Tyrosine phosphatase 1B
|
|
|
- 1)
-
Formation of a vinyl ether between Ser and Cys that result in the elimination of the thiol group from Cys (a)
- 2)
-
Sulfenyl amide (S–N bridge) between Cys-OH and main-chain amide of Ser residue (b)
|
|
Mass spectrometry (a)
X-ray crystallography (b) |
[26][31] | [41,46] |
| Cys-Phe |
hGH |
Crosslink between thioaldehyde from Cys and dehydrophenylalanine generated from Phe |
Mass spectrometry |
[26] | [41] |
Cys-DHA
Cys-DHB |
Lens proteins (βB1, βB2, βA3, βA4 and γS crystallins) |
Nucleophilic addition from Cys (GSH) to DHA or DHB |
Mass spectrometry |
[32] | [47] |
| Tyr-Gly |
Insulin |
Michael addition of primary amines (N-terminal Gly) to oxidized Tyr species |
Mass spectrometry |
[33] | [48] |
| Trp-Gly |
Matrilysin (Matrix metalloproteinase 7) |
Crosslink between 3-chloroindolenine (3-Cl-Trp) and the main-chain amide adjacent to a Gly |
NMR spectroscopy |
[34] | [49] |
| Tyr-His |
Insulin |
Michael addition from His to oxidized Tyr |
Mass spectrometry |
[33] | [48] |
Tyr-Tyr
(selected data) |
Isolated proteins including: α-lactalbumin, caseins, glucose 6-phosphate dehydrogenase, lysozyme, fibronectin, laminins, tropoelastin, cAMP receptor protein, α-synuclein, calmodulin, insulins, hemoglobin, human Δ25 centrin 2.
Human lipoproteins
Human plasma proteins, including those from people with chronic renal failure
Human atherosclerotic lesions
Erythrocytes exposed to H | 2 | O | 2 |
Brain proteins (amyloid-beta and α-synuclein) from Alzheimer’s subjects
Lipofuscin from aged human brain
Urine from people with diabetes
Human lens proteins
Bacterial spore coat proteins
Parasite oocysts |
C–C and/or C–O crosslinks via radical–radical reactions |
Western blotting
UPLC/HPLC with various detection methods
Mass spectrometry |
[33][35][36][37][38][39][40][41][][62] | [48,50,51,52,53,54,55 | 42][43][44][45][46][47][48][49 | ,56 | ][50 | ,57 | ] | ,58,59,60,61,62 | [51][52][53][54] | ,63 | [ | ,64 | 55 | ,65,66 | ] | ,67,68 | [ | ,69 | 56] | ,70 | [57] | ,71 | [58][59][60 | ,72 | ] | ,73 | [ | ,74,75 | 61 | ,76,77] |
| Trp-Trp |
|
- a)
-
α-Lactalbumin
- b)
-
Superoxide dismutase 1 (hSOD)
- c)
-
Lysozyme-hSOD
- d)
-
αB-Crystallin
- e)
-
Fibronectin
|
|
C–C or C–N crosslinks via radical–radical reactions |
Mass spectrometry |
[35][42]] | [50 | [63 | ,57 | ][64][65 | ,78,79,80] |
| Tyr-Trp |
|
- a)
-
Cytochrome c peroxidase
- b)
-
α-Lactalbumin
- c)
-
Glucose 6-phosphate dehydrogenase
- d)
-
Lysozyme
- e)
-
β-Crystallin
- f)
-
Human cataractous lenses
- g)
-
Fibronectin
|
|
C–C (or C–O and C–N) crosslinks via radical–radical reactions |
X-ray crystallography (a)
Mass spectrometry (b–g) |
[35][38][41][42][65][66] | [50,53,56,57,80,81] |
| His-His |
|
- a)
-
Immunoglobulin G1
- b)
-
Immunoglobulin G4
- c)
-
N-Ac-His
|
|
Nucleophilic addition of His to oxidized His |
Mass spectrometry (a,b)
NMR (c) |
[67][68][69][70] | [82,83,84,85] |
| His-Arg |
Ribonuclease A (RNAse) |
Nucleophilic addition of Arg to oxidized His |
Mass spectrometry |
[71] | [86] |
| His-Lys |
Immunoglobulin G1 |
Nucleophilic addition of Lys to oxidized His |
Mass spectrometry |
[67][69] | [82,84] |
| His-Cys |
Immunoglobulin G1 |
Nucleophilic addition of Cys to oxidized His |
Mass spectrometry |
[69] | [84] |
| Tyr-Lys |
|
- a)
-
RNAse
- b)
-
Interferon beta-1a
- c)
-
Insulin
|
|
Michael addition of Lys to oxidized Tyr |
Mass spectrometry |
[33][71][72] | [48,86,87] |
67. Secondary Reactions of Crosslinks
In most biological systems, protein crosslinks, and particularly the formation of irreversible covalent crosslinks such as di-Tyr, di-Trp and Tyr–Trp, are considered as ‘final’ oxidation products
[73][181]. However, over-oxidation of these species is possible, particularly under conditions of extensive oxidative damage, or under environments with long-term protein exposure to oxidants, where secondary one-electron oxidation with formation of radicals such as di-Tyr
•, di-Trp
•, or Tyr–Trp
• may occur. Such radicals can mediate similar reactions to those described above for Tyr
• and Trp
•, including reaction with O
2 to produce oxygenated products (e.g., alcohols and hydroperoxides) and self-reactions to generate trimers and oligomers. Thus, formation of tri-Tyr and pulcherosine crosslinks have been detected in human phagocytes
[13][19], while di-, tri- and tetra-Tyr have been reported in structural proteins of plant parasitic nematodes
[74][182]. In addition, oligomers of Tyr (
n = 2–8) have been reported in α-lactalbumin exposed to a horseradish peroxidase–H
2O
2 system
[75][183]. Tri-Trp has been reported in trimers of hSOD1 triggered by CO
3•− [63][78], while tri-Trp and a di-Trp hydroperoxide (di-Trp-OOH) were reported in solutions of free Trp and riboflavin illuminated with a high-intensity 365 nm light-emitting diode
[76][162].
In contrast, photo-oxidation (at 320 nm) of di-Tyr, in the presence of O
2, has been reported to occur via processes involving O
2•−, singlet oxygen (
1O
2) and H
2O
2 [77][184]. These observations were ascribed to the action of di-Tyr crosslinks as photosensitizers that could induce photo-damage to other biomolecules
[77][184]. These findings suggest that di-Tyr crosslinks may be able to extend oxidation processes, opening new pathways of reactions, though these are only likely to be of major impact in systems with very extensive extents of oxidation. The scope and role of these pathways is unexplored, as well as the ability of peroxides such as di-Trp-OOH to extend protein oxidation (in line with the capacity of other hydroperoxides
[16][31]).
78. Detection of Crosslinks, including Advantages and Disadvantages of Different Methods
7.1. Analysis of Changes in Molecular Mass by Electrophoresis and Size Exclusion Chromatography (SEC)
8.1. Analysis of Changes in Molecular Mass by Electrophoresis and Size Exclusion Chromatography (SEC)
Protein electrophoresis (e.g., SDS-PAGE) and SEC-derived methodologies are excellent tools to assess the presence of crosslinked proteins in samples. Separation by electrophoresis is typically achieved through the use of polyacrylamide gels with different pore sizes. Diverse strategies can be utilized with this approach, such as running gels under native, denaturing and/or reducing conditions. This allows the investigation and differentiation of the contributions of reducible intermolecular bonds (e.g., disulfides between two protein chains) from non-reducible bonds (e.g., carbon–carbon bonds).
Whilst these techniques can provide information on the presence of
intermolecular bonds, they rarely provide information about the presence of
intramolecular species. Moreover, data analysis needs to be carried out with care, as multiple proteins may be present in each band/fraction from complex matrices.
7.2. Analysis of Protein Crosslinks by Western (Immuno-) Blotting and ELISA Assays
8.2. Analysis of Protein Crosslinks by Western (Immuno-) Blotting and ELISA Assays
The use of antibodies to investigate the formation of oxidation products, including di-Tyr, is a widely used strategy. These are typically examined using immunoblotting or ELISA assays, with the former providing (limited) information on the nature and identity of the proteins on which the crosslinks are present, and whether these are intramolecular (in a monomer) or interchain species. However, there are few well-characterized antibodies against crosslinked species, and these vary significantly in their specificity and selectivity, with some having significant cross-reactivity with other materials. Furthermore, crosslinks buried within highly aggregated species may be poorly, or not, recognized by (large) antibodies. Thus, appropriate control experiments are critical, and both positive and negative data should be validated by alternative methods.
7.3. Direct Detection by Spectrophotometric and Fluorometric Assays
8.3. Direct Detection by Spectrophotometric and Fluorometric Assays
Some crosslinked species, as well as heavily aggregated proteins, can be monitored by spectrophotometry and fluorescence spectroscopy. For example, turbidity changes in solutions can be used (in an approximate manner) to monitor the time-course of protein crosslinking and aggregation
[78][79][189,190]. However, this approach does not provide information on the type and nature of the crosslinked proteins and is useful only when there is extensive formation of heavily aggregated species, as soluble dimers and oligomers do not contribute significantly to the turbidity of solutions.
In contrast, fluorescence experiments can provide relatively specific data on crosslinks such as di-Tyr (excitation and emission maximum at ~280 and ~410 nm, respectively)
[80][191]. Such data need to be interpreted with care, particularly with intact proteins or complex systems, as other fluorescent or optically absorbing species (e.g., Tyr, Trp, Trp-derived products, co-factors) may be present that distort excitation or emission processes.
7.4. HPLC/UPLC Methodologies
8.4. HPLC/UPLC Methodologies
These techniques can provide important quantitative data on both the consumption of the parent amino acid residues, and product formation, including Trp- and Tyr-derived crosslinks
[81][80][82][83][158,191,193,194].
7.5. Detection and Characterization of Crosslinked Proteins Using Other Biophysical Approaches
8.5. Detection and Characterization of Crosslinked Proteins Using Other Biophysical Approaches
Biophysical techniques including circular dichroism (CD), light scattering, small angle neutron scattering (SANS), small angle X-ray scattering (SAXS), X-ray crystallography, NMR spectroscopy, and electron microscopy can provide useful information on protein structure. These methods are sensitive to modified structures, supplying valuable information on changes on morphology (i.e., mass, size and shape), secondary structure and solubility
[30][84][85][45,196,197]. Some of these can also yield data on increased electron density between residues, thus supporting the presence of both intra- and intermolecular crosslinked species. This approach has been used in X-ray crystallographic studies, to determine the exact sites of crosslinks in oxidized peroxiredoxin 5, thioredoxin 2 and γS-crystallin, and to elucidate a covalent crosslink between Cys and Lys containing an N–O–S bridge
[30][85][86][87][45,197,198,199]. However, most of these methods (with the exception of X-ray crystallography, NMR spectroscopy and cryogenic electron microscopy) cannot provide a structure of sufficiently high-resolution to provide definitive identifications and they must therefore be combined with other methodologies. Moreover, these methods are currently limited to homogeneous (single protein) samples that are available in large quantities (mg amounts).
7.6. Mass Spectrometry (MS)-Based Detection and Structural Characterization of Crosslinked Proteins
8.6. Mass Spectrometry (MS)-Based Detection and Structural Characterization of Crosslinked Proteins
MS is a highly versatile technique for analysis of protein crosslinks that can be applied to (i) detect crosslinks and quantify their abundance, (ii) localize the specific crosslinking sites within polypeptides and (iii) reveal the identity of the crosslinked proteins. All of these questions cannot, however, be readily answered in a single experiment, and careful consideration must be given to appropriate workflows for specific applications.
89. Crosslink Quantification
At present, there are few methods that allow absolute quantification of crosslink concentrations, with a major limitation being the non-availability of pure standards, particularly from commercial sources. Thus, there is a pressing need for further pure crosslink standards for quantitative analyses. Disulfides are a major exception, together with di-Tyr (which is commercially available) and a few species generated via glycation reactions (e.g., pentosidine). Di-Tyr can therefore be quantified in absolute terms using some of the methods outlined above, using the purified material to construct standard curves (e.g., for MS or fluorescence detection, and UPLC/LC separation). Relative concentrations can also be obtained from ELISA assays using an anti-di-Tyr antibody, and approximate concentrations from MS analyses. In nearly all cases, even with isolated proteins where protein turnover is not a confounding factor, the absolute concentrations determined by these approaches give low values, suggesting that there are many other important crosslinks which are either undetected, or
which we cannot quantify.